Abstract
Lipid nanodiscs provide a native-like lipid environment for membrane proteins and they have become a valuable platform for the study of membrane biophysics. A range of biophysical and biochemical analyses are enabled when membrane proteins are captured in lipid nanodiscs. Two parameters that can be controlled when capturing membrane proteins in lipid nanodiscs are the radius, and hence the surface area of the lipid surface, and the composition of the lipid bilayer. Despite their emergence as a versatile tool, most studies with lipid nanodiscs in the literature have focused on nanodiscs of a single radius with a single lipid. In light of the complexity of biological membranes it is likely that nanodiscs with multiple membrane components would be more sophisticated models for membrane research. It is possible to prepare nanodiscs with more complex lipid mixtures to probe the effects of lipid composition on several aspects of membrane biochemistry. Detailed protocols are described here for the preparation of nanodiscs with mixtures of phospholipids, incorporation of cholesterol, and incorporation of a spectroscopic lipid probe. These protocols provide starting points for the construction of nanodiscs with more physiological membrane compositions or with useful biophysical probes.
Keywords: lipid nanodiscs, mixed lipids, laurdan, cholesterol, membrane protein
INTRODUCTION
Lipid nanodiscs have revolutionized membrane biochemistry by enabling a wide range of biophysical methods to study membrane properties and to better understand the structure and function of membrane proteins. Among the various nanodisc platforms that have been developed, including bicelles and polymer-based nanodiscs (Dorr et al., 2016; Durr, Gildenberg, & Ramamoorthy, 2012; Ravula, Hardin, Di Mauro, & Ramamoorthy, 2018), the protocols described here are focused on lipid nanodiscs that include engineered Apolipoprotein A1. Sligar and co-workers originally envisioned the potential utility of these lipid nanodiscs to capture membrane proteins in a ‘model’ lipid bilayer and thus provide a mechanism to stabilize and monomerize proteins that were historically difficult to purify and characterize (Civjan, Bayburt, Schuler, & Sligar, 2003; Denisov, Grinkova, Lazarides, & Sligar, 2004). They performed extensive optimization for the preparation of nanodiscs, including different lipids for reconstitution, and they performed detailed characterization of the resulting particles. In addition, they engineered the apolipoprotein structure, or membrane scaffold protein, in order to tune the diameter of the resulting nanodiscs. Since their initial efforts, these lipid nanodiscs have become a common platform for membrane biophysical studies. Arguably, any lab interested in membrane biophysics is likely to be aware of, and to have considered the application of, lipid nanodiscs if they have not already incorporated them into their research. Accordingly, many aspects of nanodiscs have been reviewed in detail in the literature (Denisov & Sligar, 2016, 2017).
Having endured the ‘growing pains’ of a new technology, nanodisc science is entering a phase of increased sophistication where the complexity of the lipid component has been expanded to more closely mimic the complexity of biological membranes and to explicitly probe the role of specific lipids in membrane characteristic and protein function.
As noted above, an obvious utility of mixed lipid nanodiscs is to understand the physical properties of membranes including their fluidity, lateral heterogeneity, and charge distributions. It is well established that the constituents of the membrane can influence protein function and cellular processes through ‘nonspecific’ physical properties (Her et al., 2016; Morigaki & Tanimoto, 2018; Shaw, McLean, & Sligar, 2004) but the details of these effects remain unresolved. Similarly, specific interactions between lipids including cholesterol with ‘binding sites’ on proteins contribute to the overall energetics of the folded protein and functionally relevant conformational changes (Corradi et al., 2019; Fantini & Barrantes, 2013; Fantini, Epand, & Barrantes, 2019; Kimura, Jennings, & Epand, 2016). As our appreciation for the complexity and sophistication of membrane-protein interactions increases, it is likely that interest in nanodiscs with greater complexity and sophistication will increase in parallel. Therefore, specific protocols to generate nanodiscs with lipids mixtures is warranted.
From our experience, it appears that generation of nanodiscs with simple binary lipid mixtures is relatively straightforward (Fig. 1), and we expect that the general procedures outlined here will be good starting points for generating nanodiscs with other lipids.
Figure 1.
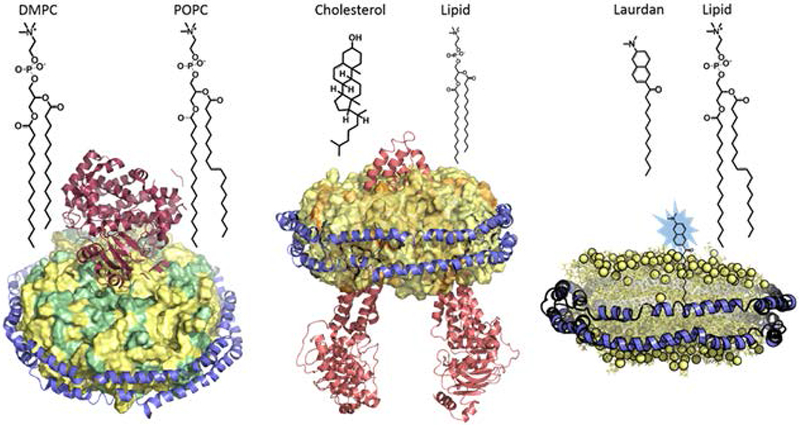
Nanodiscs with lipid mixtures. Reconstitution of mixed lipid nanodiscs containing two lipids (colored green and yellow) with different acyl chains and saturation is described in basic protocol 1 (left), lipid nanodiscs containing cholesterol (colored orange) is described in basic protocol 2 (middle), and nanodiscs containing the fluorescent probe laurdan (colored blue) is described in basic protocol 3 (right).
With that general perspective, however, it is important to note some remaining uncertainty. For the generation of nanodiscs with lipid mixtures, it is routine to prepare lipid films with a defined stoichiometry of lipids, and this can be done precisely. However, in subsequent steps that rely on the self-assembly of nanodiscs from lipid mixtures and membrane scaffold proteins (MSPs), it is assumed that the final lipid nanodiscs incorporate a lipid ratio equivalent to that of the initially prepared lipid films, but this is not typically validated experimentally. The available experimental data from characterization of various properties of nanodiscs made from lipid mixtures clearly indicate that they are different from nanodiscs made from single lipid components. So, it is clear that some mixture of lipids is incorporated in these nanodiscs. Analytical methods for validation of the exact lipid content of the nanodiscs after self-assembly from individual components are not well established and this, arguably, is a critical issue to deal with in the future.
STRATEGIC PLANNING
Selection of lipid
Lipid composition affects membrane properties as well as protein structure, function and dynamics, therefore the careful selection of an appropriate lipid can greatly benefit protein reconstitution and biophysical studies. A preliminary step is to consider the native lipid environment of the protein by performing a literature search, or a lipid screen with activity assays (if available) to select a lipid that does not negatively impact the functional activity or stability of the protein. As a start, phosphatidylcholine (PC) is known to be a major component of the plasma membrane lipid bilayer, and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) is one of the most widely utilized lipids in nanodisc reconstitution. Alternative lipids include synthetic PC lipids such as 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) or 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC).
Another important consideration is the lipid requirement or preference of a protein target. In cases where protein targets are shown to have a strong preference for anionic lipids for functional activity, anionic lipids such as 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE)/1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-Dimyristoyl-sn-glycero-3-phosphorylglycerol (DMPG)/1,2-Dioleoylphosphatidylglycerol (DOPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (POPS) and 1,2-dioleoyl-sn-glycero-3-phosphate (DOPA) have been used for nanodisc reconstitution. As a note of caution however, due to the charge properties of these lipids, low protein incorporation into nanodiscs has been reported for DOPA and DOPE (Rues, Dotsch, & Bernhard, 2016).
Once a membrane target is successfully incorporated into nanodiscs containing a single lipid, more complex lipid nanodiscs can be attempted by the addition of other lipids and/or cholesterol in ratios that resemble the plasma membrane or alter membrane fluidity. Certain membrane proteins incorporate preferentially into nanodiscs of specific lipid composition. For example, GPCRs are stabilized by cholesterol and preferentially incorporate into lipid nanodiscs containing cholesterol (Bocquet et al., 2015). Similarly, proteins that are associated with lipid rafts may require lipid environments that are enriched with cholesterol and glycosphingolipids.
Sometimes, natural lipid extracts (e.g. E. coli polar or total lipid extract) containing a mixture of lipids are utilized for nanodisc reconstitution, particularly if the membrane protein has been demonstrated to be functionally active in these lipid extracts (Finkenwirth et al., 2015). In a few cases, protein-incorporated nanodiscs are assembled directly from cell membranes or native tissue membranes (Gregersen, Fedosova, Nissen, & Boesen, 2016). While this method minimizes the exposure of membrane proteins to detergents during extensive purification steps, large amounts of MSP need to be added at the onset of cell membrane solubilization and the yield of target protein incorporated into nanodiscs is typically low after separation from nanodiscs containing non-target proteins.
In summary, selecting a suitable lipid composition for nanodiscs reconstitution depends on the goal/nature of the experiment and functional properties of the protein. One basic consideration at any point is the thermal stability of the protein target. If the protein is unstable or rapidly degrades at higher temperatures, lipids with higher phase transition temperature (e.g. DPPC, Tm ~ 41°C) may be unsuitable due to prolonged incubations at these temperatures for nanodisc assembly. In such cases POPC may be more favorable due to a low phase transition temperature at ~4°C. Lipid phase transition temperatures are typically provided by Avanti Polar Lipids, Inc. (Alabaster, AL, USA), however, phase transition temperatures can be experimentally determined calorimetrically (Biltonen & Lichtenberg, 1993) or, alternatively, using light scattering measurements (Michel, Fabiano, Polidori, Jack, & Pucci, 2006). If the experimental goal is to reconstitute a novel membrane protein or work with a known protein with limited information, and it has been demonstrated that the protein is functionally active in a single lipid composition, forming nanodiscs of a single lipid composition is recommended as a starting point. Next, if the aim is to investigate the effects of membrane fluidity, to study a protein in a more native-like environment, or to improve incorporation of a specific membrane protein in the presence of other lipids, a mixture of two or more lipids and/or cholesterol may be considered for nanodisc reconstitution. At this stage, some factors to consider include the lipid requirement of the target protein. If a membrane protein strongly prefers anionic lipid, nanodiscs with a mixture of anionic lipids may be required. Cholesterol may be supplemented when reconstituting membrane proteins like GPCRs. Furthermore, when working with recalcitrant proteins that are difficult to purify or reconstitute into lipid membranes, direct reconstitution from cells or native tissue membranes sometimes offer a viable solution. After nanodiscs reconstitution, if subsequent biophysical experiments require the monitoring of temperature-dependent transitions, for example thermal shift assays such as differential scanning calorimetry (DSC), a lipid with a specific transition temperature may be chosen favorably by considering the lipid contribution to a thermogram and to minimize overlap with the protein spectral peak.
Choice of solubilizing detergent
Detergents are not only important in the solubilization and purification of membrane proteins, but also in nanodisc assembly by ensuring all components (lipids and proteins) are well solubilized. Non-ionic detergents are considered to be relatively mild and the least denaturing among all the classes of detergents, and alkyglucosides such as n-dodecyl-β-D-maltoside (DDM) are often the best choice for successful reconstitution of functional proteins. Shorter chain non-ionic detergents such as octyl-β-glucoside (OG) are also widely used for membrane protein reconstitution but may lead to deactivation of the protein unlike the longer chain derivatives (Seddon, Curnow, & Booth, 2004). While most detergents require extensive dialysis or long incubations with adsorbent polymer beads for complete removal, using OG when it is non-detrimental to the protein has the benefit of relatively fast and efficient removal by dialysis or gel filtration. Although the initial protocols of nanodisc reconstitution recommend cholate as the solubilizing detergent, detergents including Triton X-100 and 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) have been used with success. Combinations of more than one detergent are increasingly adopted to solubilize difficult membrane targets followed by nanodisc reconstitution. For example, 2,2-didecylpropane-1,3-bis-β-D-maltopyranoside (LMNG) is gaining popularity as a very mild detergent (very low CMC value) for solubilizing difficult membrane targets and retaining protein function, and nanodisc reconstitution has been performed by adding LMNG-purified protein to cholate- or Triton X-100-solubilized mixture of lipids and MSP (Kraft, Hresko, & Hruz, 2015).
Empirical optimization of lipid:MSP ratio for each individual lipid mixture
Optimizing the ratio of lipid-to-MSP during assembly ensures the formation of a monodisperse population of lipid nanodiscs. If the ratio is too large, polydisperse particles resembling liposomes or larger aggregates tend to form and may present as a slightly turbid solution. On the other hand, if the ratio is too small, smaller nanoparticles that are lipid-scarce are formed, usually accompanied by the presence of free MSP as detected by size-exclusion chromatography (SEC). A ratio that is optimal results in a monodisperse peak at the correct elution time corresponding to the nanodiscs size during SEC, without any aggregation or large particles or free scaffold proteins. Depending on the lipid, MSP, detergent and buffer conditions, the lipid to MSP ratio varies between protocols. The ratio may also vary slightly within a protocol, depending on the accuracy of concentrations measured for each component (i.e. lipid or MSP concentration); therefore, determining the optimal lipid:MSP ratio for each batch of nanodiscs is recommended, for example, with a new batch of MSP or lipid stock. The lipid-to-MSP1D1 ratios for single lipid and lipid mixtures listed in Table 1 (from both lab-optimized procedures and published protocols) thus serve as a reference for determining the optimal ratio.
Table 1.
Optimized Lipid to MSP1D1 Ratios
Verifying the assembly of nanodiscs
At the end of nanodisc assembly, it is necessary to determine the quality of the assembled nanodisc product. In addition to the observation of a monodisperse peak corresponding to the nanodisc size during SEC, nanodisc formation can be determined using negative-stained electron microscopy. Protein incorporation into nanodiscs has also been quantitated using GFP-tagged proteins (Rues et al., 2016). To measure changes in membrane fluidity upon forming nanodiscs with lipid mixtures, the fluorescent probe laurdan can be incorporated during nanodisc assembly (Denisov, McLean, Shaw, Grinkova, & Sligar, 2005; McClary, Sumida, Scian, Paco, & Atkins, 2016). Finally, the combination of biochemical assays to measure protein activity post-nanodiscs formation and biophysical methods (such as DSC) to probe transition temperature changes of the protein or lipid mixture will help to inform on the success of nanodisc reconstitution.
The protocols in this article describe the step-by-step methods to generate nanodiscs with 2 lipids (Basic Protocol 1), nanodiscs with cholesterol (Basic Protocol 2) and fluorescence polarization measurements with laurdan (Basic Protocol 3). While these steps describe nanodisc formation with MSP1D1, the same steps would apply to forming nanodiscs of other sizes (such as with MSP1E3D1) but with different optimal lipid: scaffold protein ratios.
BASIC PROTOCOL 1
MIXED LIPID NANODISCS
Nanodisc preparations have typically used lipids which all contain the same head group, acyl chain length, and saturation. However, because nanodiscs are formed in vitro, precise control over lipid composition can be achieved. We provide a protocol for assembling nanodiscs composed of two different lipids, allowing for the study of more complex membrane-protein interactions and/or membrane-small molecule interactions.
Materials
Disc Formation Buffer (100 mM potassium phosphate, pH 7.4, 50 mM sodium chloride)
Lipid (Powder Form, Avanti Polar Lipids)
Chloroform (≥99.5%; CAS: 67-66-3; #C2432)
Methanol (≥99.9%; CAS: 67-56-1; #34860)
200 mM Sodium Cholate Hydrate (≥99%; CAS: 206896-87-0; #C6445)
Amberlite® XAD®-2 beads (#20275)
Hamilton Syringe (cleaned with fresh chloroform; 10 µL, #HAM80075; 1mL, #22192-U)
Glass Test Tubes (cleaned with fresh chloroform; Fisher Scientific)
Nitrogen Gas
Vacuum Desiccator (e.g. Kimble-Chase)
Water Bath Sonicator (e.g. Branson)
MSP1D1 (≥90% pure; expressed and purified in-house (Ritchie et al., 2009) or can be purchased from SigmaAldrich, #M6574)
0.2 µm Syringe Filter (WHA67841302)
Superdex 200 10/300 GL column (#17517501, GE Healthcare)
HPLC instrument (e.g. Shimadzu)
NOTE: Unless otherwise stated, all materials were purchased from SigmaAldrich.
Preparation of Lipid Film
-
1
Dissolve the desired powdered lipids separately in chloroform, aiming for a desired stock concentration of 130-150 mM for each (e.g. dissolve 100 mg of POPC [MW = 760.1 g/mol] in 877 µL of chloroform to make a 150 mM stock solution).
NOTE: Use glassware only when working with organic solvents such as chloroform. Chloroform is highly volatile and toxic at high concentrations. Working in a fume hood is recommended.
-
2
Measure the lipid stock concentrations by performing a phosphate assay of the lipid-chloroform stock prepared Step 1.
NOTE: For best accuracy, the phosphate assay should be performed in both 1 µL and 2 µL triplicates. For a complete phosphate assay procedure, please refer to the ‘Determination of Total Phosphorus’ procedure described by Avanti Polar Lipids, Inc. (see Internet Resources).
-
3
Transfer the two chloroform-dissolved lipid stocks to a clean glass test tube in the desired molar ratio using a clean Hamilton syringe and briefly swirl gently to ensure complete mixing.
-
4
Remove the chloroform by blowing a gentle stream of nitrogen into the tube while rotating the tube at an angle. To facilitate removal of chloroform, the tube can be warmed during this process by pressing the bottom against a warm surface (e.g. the palm of a hand).
-
5
Place the test tube into a vacuum desiccator for a minimum of 8 hours to remove any residual chloroform while minimizing oxidation of any unsaturated lipids.
NOTE: Avoid using too strong of a stream of nitrogen gas, as this can result in splashing of the lipid away from the bottom of the tube. Chloroform is sufficiently evaporated when liquid is no longer visible at the bottom of the tube and a thin opaque film has formed on the tube wall. See Fig. 2 for a schematic diagram of chloroform removal from lipids.
Figure 2.
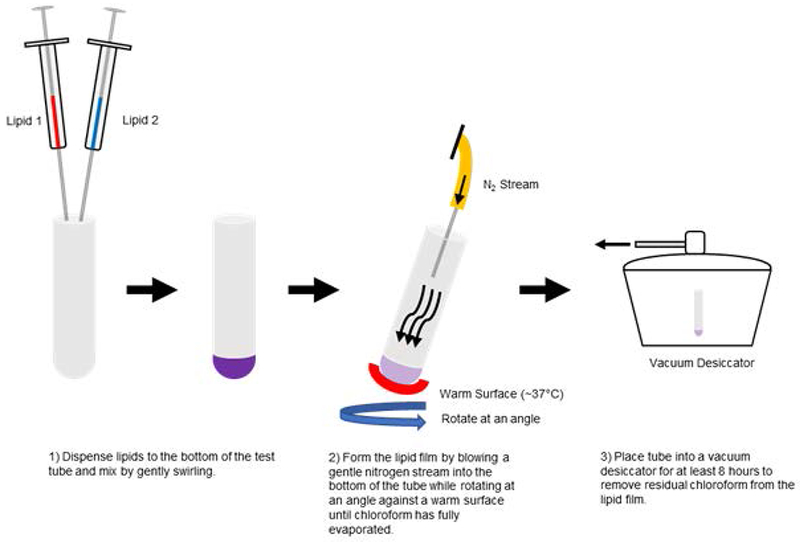
Schematic diagram describing the chloroform removal process for the preparation of lipid films. All test tubes and syringes should be cleaned by rinsing with fresh chloroform immediately prior to use.
Preparation of Amberlite® XAD®-2 Beads
-
6
Add Amberlite® XAD®-2 beads to a 50 mL conical tube up to the 25 mL line.
-
7
Add nanopure water to the tube up to 50 mL and invert several times to wash the beads.
-
8
Carefully decant the water until the settled bead level is approximately 25 mL.
-
9
Add methanol to the tube up to 50 mL and invert several times to wash the beads.
-
10
Carefully decant the methanol until the settled bead level is approximately 25 mL.
-
11
Repeat steps 9 and 10 once.
-
12
Add water to the tube up to 50 mL and invert several times to wash the beads.
-
13
Carefully decant the water until the bead level is approximately 25 mL.
-
14
Repeat steps 12 and 13 an additional 5 times.
-
15
Store beads such that they are submerged under a 10-15 mL layer of water and store at 4°C until use.
NOTE: If bacterial growth is of concern, sodium azide may be added to the washed beads up to a final concentration of 0.1%.
Nanodisc Assembly
-
16
Add a 50 mM stock solution of sodium cholate (dissolved in disc formation buffer) to a final molar ratio of 2:1 (cholate:lipid).
-
17
Cover the test tube with parafilm.
-
18
Rotate the bottom of the test tube in a water bath sonicator for 10 second intervals with 10 second rests until the lipid film is dissolved and/or homogeneous in appearance (see Troubleshooting section).
-
19
Mix MSP1D1 with dissolved lipid, disc formation buffer, and extra sodium cholate such that the final sodium cholate concentration is 20 mM (2 x the critical micelle concentration).
-
20
Incubate for 1 hour on an orbital shaker near the average transition temperature of the lipid mixture.
-
21
Using a metal spatula, take an aliquot of Amberlite® XAD®-2 beads, wick away excess liquid, and add 0.5 g beads per every 1 mL of assembly mixture.
-
22
Let the reaction incubate on an orbital shaker for an additional 4 hours.
-
23
Remove the beads by passing the assembly mixture through a 0.2 µm syringe filter.
-
24Analyze nanodiscs using a Superdex 200 10/300 GL column and monitoring absorbance at 280 nm:
- Running Buffer = Disc Formation Buffer
- Flow Rate = 0.5 mL/min
- Length of Elution = 60 minutes
- Sample injection = typically 20 to 40 ug MSP1D1 protein; injection volume should not exceed 500 µL.
BASIC PROTOCOL 2
NANODISCS WITH CHOLESTEROL
Cholesterol alters membrane fluidity and stabilizes membrane proteins such as GPCRs and transporters such as P-glycoprotein. The addition of 10-30% cholesterol strongly modifies the properties of the lipid bilayer and is commonly used for studying membranes and proteins. The following protocol describes the steps to incorporate 20% cholesterol into E. coli total lipid and DMPC nanodiscs. In general, the addition of cholesterol decreases the ratio of lipid to MSP1D1, likely because cholesterol reduces lipid packing and hence less lipid is required per disc.
Materials
Disc Formation Buffer (20 mM Tris base, pH 7.4, 150 mM sodium chloride)
Lipid (Powder form, Avanti Polar Lipids)
Cholesterol hemisuccinate (CHS) (CAS: 1510-21-0; #C6512)
100 mM n-dodecyl-β-D-maltopyranoside (nDDM) (CAS: 69227-93-6; #D310, Anatrace)
Methanol (≥99.9%; CAS: 67-56-1; #34860)
Amberlite® XAD®-2 beads (#20275)
Hamilton Syringe (cleaned with fresh chloroform; 10 µL, #HAM80075; 1mL, #22192-U)
Glass Test Tubes (cleaned with fresh chloroform; Fisher Scientific)
Nitrogen Gas
Vacuum Desiccator (e.g. Kimble-Chase)
Water Bath Sonicator (e.g. Branson)
MSP1D1 (≥90% pure; expressed and purified in-house (Ritchie et al., 2009) or can be purchased from MilliporeSigma, #M6574)
0.2 µm Syringe Filter (WHA67841302)
Superdex 200 10/300 GL column (#17517501, GE Healthcare)
HPLC instrument (e.g., Shimadzu)
NOTE: Unless otherwise stated, all materials were purchased from SigmaAldrich.
Preparation of Lipid Film
-
1
Weigh cholesterol hemisuccinate and lipid (powder form) such that there is 20% cholesterol by weight. For example, add 125 mg of CHS to 500 mg of lipid. Dissolve the mixture in chloroform to a concentration of 50 mg/mL.
NOTE: Use glassware only when working with organic solvents such as chloroform. Chloroform is highly volatile and toxic at high concentrations. Working in a fume hood is recommended.
-
2
Remove the chloroform by blowing a gentle stream of nitrogen into the tube while rotating the tube at an angle. To facilitate removal of chloroform, the tube can be warmed during this process by pressing the bottom against a warm surface (e.g. the palm of a hand).
-
3
Place the test tube into a vacuum desiccator for a minimum of 8 hours to remove any residual chloroform while preventing oxidation of the lipids.
-
4
Perform phosphate assay with 1-2uL of the lipid-chloroform stock from Step 1 to determine the number of moles of phospholipid. (Refer to ‘Determination of Total Phosphorus’ from Avanti Polar Lipids).
NOTE: Avoid using too strong of a stream of nitrogen gas, as this can result in splashing of the lipid away from the bottom of the tube. Chloroform is sufficiently evaporated when liquid is no longer visible at the bottom of the tube and a thin opaque film has formed on the tube wall. See Fig. 2 for a schematic diagram of chloroform removal from lipids.
Preparation of Amberlite® XAD®-2 Beads
-
5
Add Amberlite® XAD®-2 beads to a 50 mL conical tube up to the 25 mL line.
-
6
Add nanopure water to the tube up to 50 mL and invert several times to wash beads.
-
7
Carefully decant the water until the bead level is approximately 25 mL.
-
8
Add methanol to the tube up to 50 mL and invert several times.
-
9
Carefully decant the methanol until the bead level is approximately 25 mL.
-
10
Repeat steps 8 and 9 once.
-
11
Add nanopure water to the tube up to 50 mL and invert several times.
-
12
Carefully decant the water until the bead level is approximately 25 mL.
-
13
Repeat steps 11 and 12 an additional 5 times.
-
14
Store beads such that they are submerged under a 10-15 mL layer of water and store at 4°C until use.
Nanodisc Assembly
-
15
Add 100 mM n-DDM dissolved in disc formation buffer to lipids such that there is 6-fold molar excess of detergent to lipid. For example, to 1.6 µmol of lipid, add 9.6 µmol of n-DDM.
-
16
Cover the test tube with parafilm.
-
17
Rotate the bottom of the test tube in a water bath sonicator for 10 second intervals with 10 second rests until the lipid film is dissolved.
-
18
Mix MSP1D1 with the detergent-solubilized mixture from step 15 with a molar ratio of 40:1 (lipid:MSP). For example, to 1.6 µmol of lipid, add 40 nmol of MSP1D1. Add more disc-formation buffer and 500 µL of elution buffer (used during protein purification) to make a final volume of 2.5 ml. Ensure that the final glycerol level is less than 4% so that glycerol does not interfere with nanodisc assembly.
-
19
Incubate for 1 hour on an orbital shaker near the average transition temperature of the lipid mixture. For E. coli total lipid extract or DMPC, the incubation temperature is at room temperature.
-
20
Using a metal spatula, take an aliquot of Amberlite® XAD®-2 beads, wick away excess liquid, and add 0.6 g beads per every 1 mL of assembly mixture.
-
21
Let the reaction incubate on an orbital shaker for an additional 2 hours at room temperature.
-
22
Remove the beads by passing the assembly mixture through a 0.2 µm syringe filter.
-
23
Perform a lipid:MSP1D1 ratio scout by repeating step 16 with different ratios such as 30:1 40:1 and 50:1 until the optimal ratio is determined. See example chromatograms in ‘Anticipated Results’.
-
24Analyze nanodiscs using a Superdex 200 10/300 GL column and monitoring absorbance at 280 nm:
- Running Buffer = Disc Formation Buffer
- Flow Rate = 0.5 mL/min
- Length of Elution = 60 minutes
- Sample injection = typically 20 to 40 ug MSP1D1 protein; injection volume should not exceed 500 µL.
-
25
Once the optimal lipid: MSP1D1 ratio is determined, instead of 500 µL of elution buffer used in step 16, add an equivalent volume of purified protein to the nanodiscs reaction mixture for assembling protein-incorporated nanodiscs.
-
26
Protein-incorporated nanodiscs may be separated from empty nanodiscs mixture by further affinity tag purification if the tag used is different on membrane protein and MSP1D1 (or if using his-tagged membrane proteins, his-tag on MSP1D1 needs to be cleaved). An alternative is another round of SEC-HPLC to separate protein-incorporated nanodiscs from empty nanodiscs based on size.
BASIC PROTOCOL 3
INCORPORATION OF LAURDAN INTO NANODISCS FOR MEMBRANE FLUIDITY MEASUREMENTS
The fluorescent probe laurdan provides a simple solution for monitoring changes in the phase properties of the nanodisc lipid bilayer. Upon incorporation into lipid membranes, laurdan exhibits a phase-dependent shift in its fluorescence emissions spectrum (Parasassi, De Stasio, d’Ubaldo, & Gratton, 1990; Parasassi, De Stasio, Ravagnan, Rusch, & Gratton, 1991). The generalized polarization of laurdan can be calculated using the emissions spectrum of the nanodisc in order to quantify the relative fluidity of the nanodisc membrane (see Critical Parameters and Troubleshooting). Here we describe a basic protocol for the incorporation of laurdan into nanodiscs.
Materials
Laurdan (CAS: 74515-25-6; #D250, Thermo Fisher)
-
Temperature-controlled fluorimeter
NOTE: All other reagents are as listed in Basic Protocol 1.
Disc formation buffer (20 mM Tris Base, 150 mM Sodium Chloride, pH 7.4)
Preparation of Laurdan Film
Make a 2 mM stock of laurdan dissolved in chloroform and store protected from light at −20°C until further use.
Mix lipids in a clean glass test tube as described in Basic Protocol 1. Do NOT dry the lipid film.
-
Using a clean Hamilton syringe, transfer laurdan to the test tube containing the lipid solution such that the final total lipid:laurdan ratio is 200:1. Mix by briefly swirling gently.
NOTE: A ratio of 200:1 (total lipid:laurdan; w/w) is recommended in order to incorporate laurdan into nanodiscs while avoiding significant alterations of the intrinsic properties of the lipid bilayer.
Continue from step 2 of Basic Protocol 1 and follow to completion.
To determine laurdan GP, excite laurdan-containing nanodiscs at 340 nm and collect fluorescence emissions from 360 nm to 600 nm using a temperature-controlled fluorimeter. Record the emissions intensities at 440 nm and 490 nm for each temperature scan collected.
- Calculate the generalized polarization (GP) of laurdan using the following equation:
where I440 and I490 are the relative fluorescence emission intensities at 440 nm and 490 nm, respectively.
COMMENTARY
Background
It is clear that the biological membrane serves an important role beyond providing cellular compartmentalization and serving as a hydrophobic environment for membrane proteins. Indeed, the effects of membrane composition on localization and enzymatic activity has been well documented. For instance, the localization of Cytochrome P450s into lipid-ordered vs disordered domains was found to be isoform dependent (Brignac-Huber, Reed, Eyer, & Backes, 2013; Park, Reed, Brignac-Huber, & Backes, 2014). Guixa-Gonzalez and co-workers have also shown that membrane cholesterol can reach the binding pocket of the adenosine A(2A) receptor (a GPCR), potentially serving as an allosteric regulator (Guixa-Gonzalez et al., 2017).
Membrane proteins have traditionally been reconstituted for biochemical and biophysical characterization using systems such as detergent micelles, liposomes, and bicelles. However, depending on the properties to be investigated, these systems could potentially have some key disadvantages. Detergent micelles, while able to solubilize most membrane proteins, cannot effectively mimic the physical and chemical properties of the lipid bilayer. Liposomes can be formed using a wide variety of lipids. However, within a single preparation, liposome size can vary significantly, which in turn can potentially affect the degree of protein self-association within the membrane. Bicelles are the most similar in structure to nanodiscs, and their formation is not dependent on the presence of a scaffolding protein. Unfortunately, bicelle formation is highly dependent on the presence of phosphatidylcholine lipids, limiting the types of lipids that can be used in this system.
By comparison, the relatively small and consistent size of nanodiscs (~10 nm in diameter for MSP1D1 nanodiscs) largely eliminates protein oligomerization, as long as the protein of interest is incorporated into the reaction mixture at sufficiently low concentrations relative to the total lipid concentration. Furthermore, the nanodisc bilayer structure is maintained by the amphipathic membrane scaffold protein in a defined size that is stable over time. The ability to use a wider variety (and complex combinations) of lipids to form nanodiscs also becomes possible.
Critical Parameters and Troubleshooting
Concentration of Lipids – It is imperative that the lipid concentrations are accurate to ensure reproducibility when optimizing the lipid:MSP1D1 ratio. Use of a phosphate assay provides the most accurate measurement of lipid concentrations and should be performed on lipid stocks within 2-3 days of the preparation of lipid films.
Assembly Incubation Temperature – For best yields, it is typically recommended to incubate the nanodisc assembly mixture at or near the lipid transition temperature. In cases where the chosen lipids have significantly different transition temperatures and precise temperature control is not possible, it is recommended to incubate assembly reactions at or near the transition temperature of the major lipid component to be incorporated into nanodiscs. If the protein of interest is unstable or rapidly degrades at higher temperatures, choosing lipids with high phase transition temperatures may not be suitable due to prolonged incubations at these temperatures for nanodiscs assembly.
Empirical Optimization of Lipid:MSP1D1 Ratio – Due to the differences in lipid packing within the nanodisc membrane, the lipid:MSP1D1 ratio will have to be empirically optimized for every lipid combination used. To achieve this, the lipid:MSP1D1 for the individual lipids should first be separately optimized. Once the optimal ratios are known, then proceed to manually adjust the lipid:MSP1D1 ratio for the mixed lipid nanodiscs, starting from the lower ratio lipid and systematically proceeding towards the higher ratio lipid. For example, the optimal POPC:MSP1D1 ratio is 65:1, while the optimal ratio for DMPC is 80:1, so for nanodiscs composed of these two lipids one could test ratios of 65:1, 70:1, 75:1, and 80:1. Optimal nanodiscs will have a peak retention time of approximately 24 minutes (12 mL) on a Superdex 200 10/300 GL column with a flow rate of 0.5 mL/min (nanodiscs can be detected by monitoring absorbance at 280 nm).
Detergent removal – Detergent removal is critical in nanodisc formation. Ensure that the detergent is mostly removed by extensive incubation with adsorbent polymer beads or dialysis. This can be verified by inspecting the SEC chromatogram and making sure that there are no major peaks towards the end of the run (> 40 mins). If necessary, increase the incubation time or the amount of adsorbent beads added up to 0.6 g/ml of nanodisc assembly mixture.
Laurdan Photosensitivity – Keep laurdan protected from light as much as possible in order to minimize photobleaching of the probe. For longer steps such as preparation of lipid films, the nanodisc assembly incubation, and purification by SEC, all tubes and columns may be covered in foil where necessary.
Glycerol – Glycerol content during nanodisc assembly should not exceed 4% as glycerol interferes with the assembly process. After nanodisc formation, 10-20% glycerol may be supplemented if the nanodiscs are to be stored at −80°C.
Anticipated Results
One of the advantages of the nanodisc platform is their well-defined size, which is determined by the length of the membrane scaffold-protein. In the case of MSP1D1, nanodiscs are approximately 10 nm in diameter (for diameters and retention times of nanodiscs formed with other commonly used MSPs, please see (Denisov et al., 2004) and (Ritchie et al., 2009)). Using a Superdex 200 10/300 GL size-exclusion column (GE Healthcare), it is established that optimal nanodiscs have a peak retention time of 24 minutes, or 12 mL (flow rate = 0.5 mL/min; running buffer: Disc Formation Buffer). Based on this ideal retention time, mixed lipid nanodiscs can then be empirically optimized for the lipid:MSP ratio to minimize the formation of under-sized nanodiscs (Fig. 3A) as well as the formation of larger aggregates (Fig. 3B). Nanodisc lipid content has been optimized when the chromatogram shows a largely homogeneous peak centered at the optimal peak retention time, with minimal shoulders appearing to the left or right of this peak.
Figure 3.
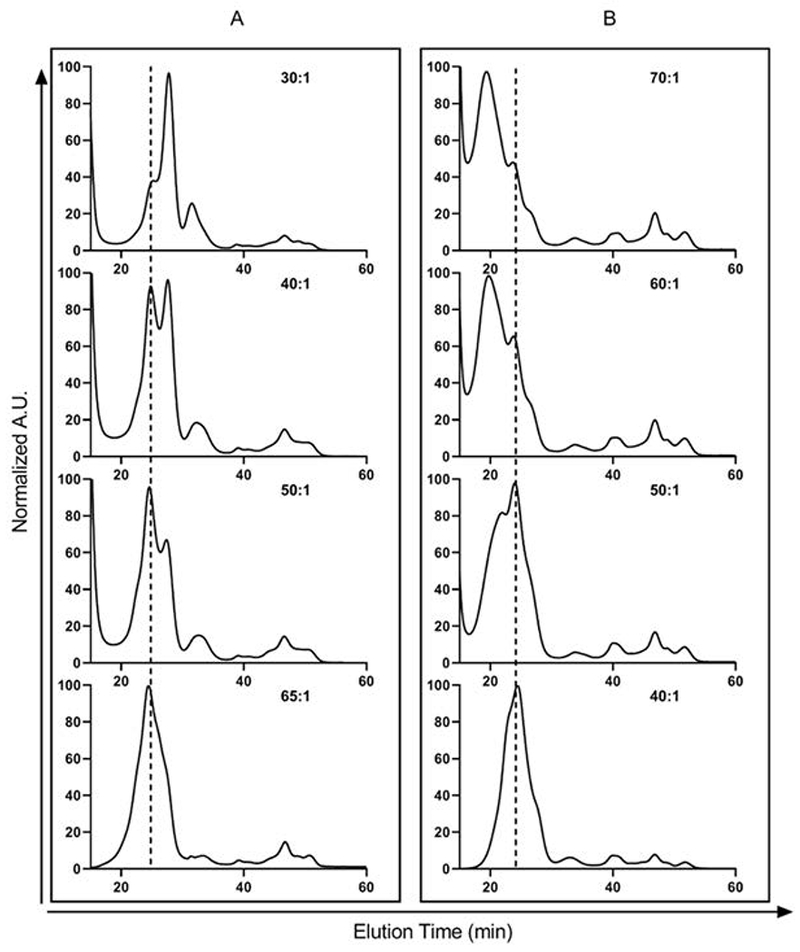
Chromatographic elution profile of mixed lipid nanodiscs (E. coli total lipid extract + 20% cholesterol) obtained by HPLC-SEC in two separate experiments (absorbance was monitored at 280 nm). (A) Starting lipid: MSP1D1 is too small, resulting in formation of smaller, sub-optimal nanoparticles (at elution time ~26 to 28 min) together with nanodiscs (~24 min). As more lipid is added (with increasing lipid:MSP1D1 ratio), a more homogeneous, monodisperse peak for nanodiscs is observed at the appropriate elution time (shown in dotted line). (B) Reverse scenario when the starting lipid:MSP1D1 ratio is too large, resulting larger aggregates eluting at or close to the void volume. Note that different batches of reagents (lipid, MSP1D1, detergent) were used for the two reconstitution trials, which resulted in different optimized lipid to MSP1D1 ratios. Relatively non-Gaussian peak profile is observed, most likely because E. coli total lipid extract is a heterogeneous mixture of phosphatidylethanolamine (PE), phosphatidylglycerol (PG), cardiolipin (CA) and other lipids.
Laurdan fluorescence is a useful tool for assessing lipid order and fluidity of the nanodisc membrane. Fluorescence emissions scans of nanodiscs containing laurdan should always show a decrease in intensity at 440 nm and an increase at 490 nm with increasing temperature (Fig. 4A) and/or increasing membrane fluidity (Fig. 4B).
Figure 4.
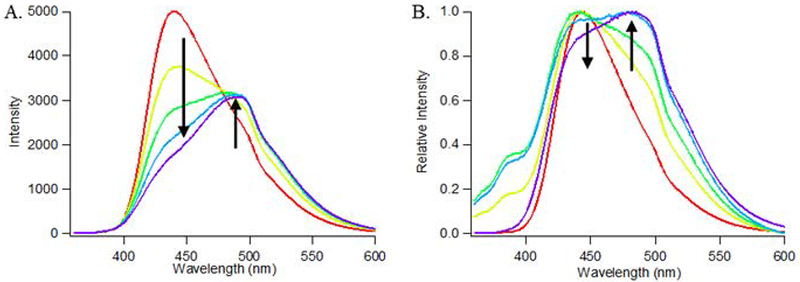
(A) Laurdan fluorescence of 100% POPC nanodiscs acquired at 5°C (red), 15°C (yellow), 25°C (green), 35°C (cyan), and 45°C (purple). A redshift in the spectra is observed at increasing temperatures (denoted by arrows). (B) Relative laurdan fluorescence of nanodiscs containing 100% DMPC (red), 80% DMPC/20% POPC (yellow), 50% DMPC/50% POPC (green), 20% DMPC/80% POPC (cyan), or 100% POPC (purple). A redshift in the spectra is observed with increasing POPC content (denoted by arrows).
Thermal melt profiles of mixed lipid nanodiscs and any incorporated membrane-bound proteins can be determined using techniques such as differential scanning calorimetry (Durowoju, Bhandal, Hu, Carpick, & Kirkitadze, 2017). Figure 5A shows the thermal melt profiles for nanodiscs of varying lipid contents. Although this method is constrained to lipid mixtures of a suitable transition temperature (> 10°C), we observe an apparent shift in the lipid transition which agrees with the previously established transition temperature of the individual lipids. Importantly, for nanodiscs containing a 1:1 ratio of POPC:DMPC, there was only a single transition temperature of ~15°C, and an absence of a DMPC transition at 24°C. This observation provides evidence that the mixed lipids are incorporated into nanodiscs and reflects the desired ratio of lipids used. However, as noted previously, this approach does not give precise measurements of lipid content per nanodisc.
Figure 5.
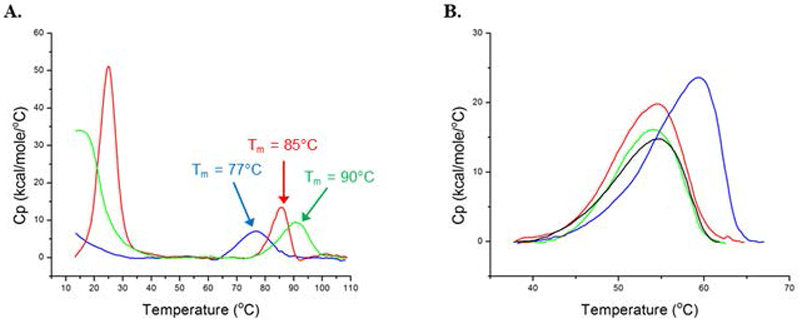
Differential scanning calorimetry of mixed lipid nanodiscs. (A) DSC thermograms of 100% POPC nanodiscs (blue), 100% DMPC nanodiscs (red), 50% POPC/50% DMPC nanodiscs (green). Lipid phase transitions all appear towards the lower temperature end of the thermogram and are consistent with the phase transition temperatures for each of the lipids (POPC Tm = −2°C; DMPC Tm = 24°C). MSP1D1 thermal unfolding transitions (denoted by arrows) are provided for reference. (B) Melt profile of CYP3A4 in 100% POPC (black), 75% POPC/25% DMPC (red), 50% POPC/50% DMPC (blue), and 25% POPC/75% DMPC (green) mixed lipid nanodiscs. [Reprinted (adapted) with permission from “Membrane Fluidity Modulates Thermal Stability and Ligand Binding of Cytochrome P450 3A4 in Lipid Nanodiscs,” by W.D. McClary, J.P Sumida, M. Scian, L. Paco, and W.M. Atkins, 2016, Biochemistry, 55, p. 6258-6268. Copyright 2016 American Chemical Society.]
Incorporation of enzymes into mixed lipid nanodiscs can alter their biochemical and biophysical properties compared to insertion of the same enzymes into nanodiscs containing a single lipid type. For example, inserting cytochrome P450 3A4 (CYP3A4) into mixed lipid nanodiscs revealed lipid-dependent differences in the thermal stability of the enzyme (Fig. 5B). When using nanodiscs containing a mixture 50% POPC/50% DMPC, CYP3A4 exhibited an increase in thermal stability, which also corresponded with an increase in the rate of binding of ketoconazole to the CYP3A4 active site (McClary et al., 2016). In another example, P-glycoprotein (P-gp) was incorporated into nanodiscs containing DMPC and 20% cholesterol, or DMPC-alone, in order to monitor the effect of cholesterol on P-gp activity (Fig. 6, A and B). Generally, the addition of 20% cholesterol to P-gp incorporated nanodiscs resulted in an approximate three-fold increase in the basal ATPase activity of P-gp. The fold stimulation of drug-stimulated ATPase activity over basal activity was lower in P-gp nanodiscs containing cholesterol (1.8 and 1.3 for verapamil and vinblastine respectively) compared to those without cholesterol (6.8 and 3 for verapamil and vinblastine respectively). This suggests that cholesterol promotes a more catalytically active state of P-gp, and thus the addition of drugs has a smaller effect in further stimulating the activity of P-gp. It is apparent that utilizing more complex membrane compositions in nanodiscs enables more detailed studies of membrane-protein interactions.
Figure 6.
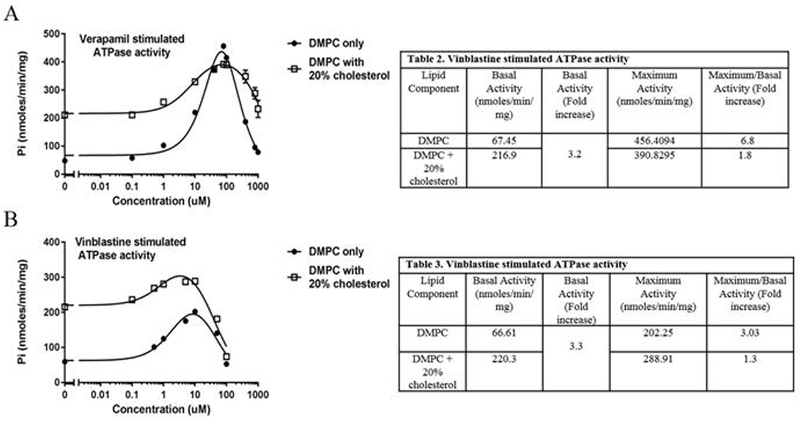
Drug-dependent ATPase activity of P-glycoprotein measured by release of inorganic phosphate, following method as described (Li, Nath, & Atkins, 2017). A) Verapamil stimulated ATPase activity of P-gp in DMPC only (black circle) or DMPC + 20% cholesterol nanodiscs (white squares). (B) Vinblastine stimulated ATPase activity of P-gp in DMPC only (black circles) or DMPC + 20% cholesterol nanodiscs (white squares).
Time Considerations
Generally, it takes anywhere from 3-4 days to express and purify MSP1D1 (Bayburt, Grinkova, & Sligar, 2002). Depending on the protein of interest and the type of purification strategies involved (i.e. affinity tag (single or dual) purification, ion-exchange chromatography, SEC), it may take anywhere from a few days to over a week for protein purification.
Making lipid films and determining the total phosphorus for a lipid stock should take a day to complete. Preparing the amberlite beads can done within the same day.
With all reagents available for nanodisc assembly, it typically takes 1-2 days to determine the optimal lipid:MSP1D1 ratio, followed by another day for incorporating the protein of interest into nanodiscs.
Measuring laurdan fluorescence for generalized polarization calculations is primarily dependent on the number of samples to be analyzed and the sample incubation temperature. As a general rule of thumb, any spectroscopic measurements collected for samples at relatively low temperatures (< 15°C) should be allowed a minimum of 10 minutes to equilibrate at the desired temperature prior to data collection. When accounting for equilibration times, a typical temperature scan experiment for laurdan-containing nanodiscs from 5 – 85°C (with 10°C increments) is expected to take approximately 1 ½ hours.
Footnotes
INTERNET RESOURCES
https://avantilipids.com/tech-support/analytical-procedures/determination-of-total-phosphorus
Details the procedure for determining total phosphorous concentration for lipid quantification.
LITERATURE CITED
- Bayburt TH, Grinkova YV, & Sligar SG (2002). Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters, 2(8), 853–856. doi: 10.1021/nl025623k [DOI] [Google Scholar]
- Biltonen RL, & Lichtenberg D (1993). The Use of Differential Scanning Calorimetry as a Tool to Characterize Liposome Preparations. Chemistry and Physics of Lipids, 64(1–3), 129–142. doi:Doi 10.1016/0009-3084(93)90062-8 [DOI] [Google Scholar]
- Bocquet N, Kohler J, Hug MN, Kusznir EA, Rufer AC, Dawson RJ, . . . Huber S. (2015). Real-time monitoring of binding events on a thermostabilized human A2A receptor embedded in a lipid bilayer by surface plasmon resonance. Biochim Biophys Acta, 1848(5), 1224–1233. doi: 10.1016/j.bbamem.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Brignac-Huber LM, Reed JR, Eyer MK, & Backes WL (2013). Relationship between CYP1A2 Localization and Lipid Microdomain Formation as a Function of Lipid Composition. Drug Metabolism and Disposition, 41(11), 1896–1905. doi: 10.1124/dmd.113.053611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civjan NR, Bayburt TH, Schuler MA, & Sligar SG (2003). Direct solubilization of heterologously expressed membrane proteins by incorporation into nanoscale lipid bilayers. Biotechniques, 35(3), 556-+. doi:DOI 10.2144/03353rr02 [DOI] [PubMed] [Google Scholar]
- Corradi V, Sejdiu BI, Mesa-Galloso H, Abdizadeh H, Noskov SY, Marrink SJ, & Tieleman DP (2019). Emerging Diversity in Lipid-Protein Interactions. Chem Rev, 119(9), 5775–5848. doi: 10.1021/acs.chemrev.8b00451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, & Sligar SG (2004). Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc, 126(11), 3477–3487. doi: 10.1021/ja0393574 [DOI] [PubMed] [Google Scholar]
- Denisov IG, McLean MA, Shaw AW, Grinkova YV, & Sligar SG (2005). Thermotropic phase transition in soluble nanoscale lipid bilayers. Journal of Physical Chemistry B, 109(32), 15580–15588. doi: 10.1021/jp051385g [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, & Sligar SG (2016). Nanodiscs for structural and functional studies of membrane proteins. Nat Struct Mol Biol, 23(6), 481–486. doi: 10.1038/nsmb.3195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denisov IG, & Sligar SG (2017). Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev, 117(6), 4669–4713. doi: 10.1021/acs.chemrev.6b00690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schafer M, van Walree CA, & Killian JA (2016). The styrene-maleic acid copolymer: a versatile tool in membrane research. Eur Biophys J, 45(1), 3–21. doi: 10.1007/s00249-015-1093-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durowoju IB, Bhandal KS, Hu J, Carpick B, & Kirkitadze M (2017). Differential Scanning Calorimetry - A Method for Assessing the Thermal Stability and Conformation of Protein Antigen. J Vis Exp(121). doi: 10.3791/55262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr UH, Gildenberg M, & Ramamoorthy A (2012). The magic of bicelles lights up membrane protein structure. Chem Rev, 112(11), 6054–6074. doi: 10.1021/cr300061w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J, & Barrantes FJ (2013). How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Frontiers in Physiology, 4. doi:ARTN 31 10.3389/fphys.2013.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantini J, Epand RM, & Barrantes FJ (2019). Cholesterol-Recognition Motifs in Membrane Proteins. Adv Exp Med Biol, 1135, 3–25. doi: 10.1007/978-3-030-14265-0_1 [DOI] [PubMed] [Google Scholar]
- Finkenwirth F, Sippach M, Landmesser H, Kirsch F, Ogienko A, Grunzel M, . . . Eitinger T. (2015). ATP-dependent Conformational Changes Trigger Substrate Capture and Release by an ECF-type Biotin Transporter. J Biol Chem, 290(27), 16929–16942. doi: 10.1074/jbc.M115.654343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen JL, Fedosova NU, Nissen P, & Boesen T (2016). Reconstitution of Na(+),K(+)-ATPase in Nanodiscs. Methods Mol Biol, 1377, 403–409. doi: 10.1007/978-1-4939-3179-8_36 [DOI] [PubMed] [Google Scholar]
- Guixa-Gonzalez R, Albasanz JL, Rodriguez-Espigares I, Pastor M, Sanz F, Marti-Solano M, . . . Selent J. (2017). Membrane cholesterol access into a G-protein-coupled receptor. Nature Communications, 8. doi:ARTN 14505 10.1038/ncomms14505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Her C, Filoti DI, McLean MA, Sligar SG, Alexander Ross JB, Steele H, & Laue TM (2016). The Charge Properties of Phospholipid Nanodiscs. Biophys J, 111(5), 989–998. doi: 10.1016/j.bpj.2016.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Jennings W, & Epand RM (2016). Roles of specific lipid species in the cell and their molecular mechanism. Progress in Lipid Research, 62, 75–92. doi: 10.1016/j.plipres.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Kraft TE, Hresko RC, & Hruz PW (2015). Expression, purification, and functional characterization of the insulin-responsive facilitative glucose transporter GLUT4. Protein Science, 24(12), 2008–2019. doi: 10.1002/pro.2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MJ, Nath A, & Atkins WM (2017). Differential Coupling of Binding, ATP Hydrolysis, and Transport of Fluorescent Probes with P-Glycoprotein in Lipid Nanodiscs. Biochemistry, 56(19), 2506–2517. doi: 10.1021/acs.biochem.6b01245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClary WD, Sumida JP, Scian M, Paco L, & Atkins WM (2016). Membrane Fluidity Modulates Thermal Stability and Ligand Binding of Cytochrome P4503A4 in Lipid Nanodiscs. Biochemistry, 55(45), 6258–6268. doi: 10.1021/acs.biochem.6b00715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel N, Fabiano AS, Polidori A, Jack R, & Pucci B (2006). Determination of phase transition temperatures of lipids by light scattering. Chem Phys Lipids, 139(1), 11–19. doi: 10.1016/j.chemphyslip.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Morigaki K, & Tanimoto Y (2018). Evolution and development of model membranes for physicochemical and functional studies of the membrane lateral heterogeneity. Biochim Biophys Acta Biomembr, 1860(10), 2012–2017. doi: 10.1016/j.bbamem.2018.03.010 [DOI] [PubMed] [Google Scholar]
- Parasassi T, De Stasio G, d’Ubaldo A, & Gratton E (1990). Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophys J, 57(6), 1179–1186. doi: 10.1016/S0006-3495(90)82637-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parasassi T, De Stasio G, Ravagnan G, Rusch RM, & Gratton E (1991). Quantitation of lipid phases in phospholipid vesicles by the generalized polarization of Laurdan fluorescence. Biophys J, 60(1), 179–189. doi: 10.1016/S0006-3495(91)82041-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, Reed JR, Brignac-Huber LM, & Backes WL (2014). Cytochrome P450 system proteins reside in different regions of the endoplasmic reticulum. Biochem J, 464(2), 241–249. doi: 10.1042/BJ20140787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravula T, Hardin NZ, Di Mauro GM, & Ramamoorthy A (2018). Styrene maleic acid derivates to enhance the applications of bio-inspired polymer based lipid-nanodiscs. Eur Polym J, 108, 597–602. doi: 10.1016/j.eurpolymj.2018.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, & Sligar SG (2009). Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. Methods in Enzymology; Liposomes, Pt F, 464, 211–231. doi: 10.1016/S0076-6879(09)64011-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rues RB, Dotsch V, & Bernhard F (2016). Co-translational formation and pharmacological characterization of beta1-adrenergic receptor/nanodisc complexes with different lipid environments. Biochimica Et Biophysica Acta, 1858(6), 1306–1316. doi: 10.1016/j.bbamem.2016.02.031 [DOI] [PubMed] [Google Scholar]
- Seddon AM, Curnow P, & Booth PJ (2004). Membrane proteins, lipids and detergents: not just a soap opera. Biochimica Et Biophysica Acta-Biomembranes, 1666(1–2), 105–117. doi: 10.1016/j.bbamem.2004.04.011 [DOI] [PubMed] [Google Scholar]
- Shaw AW, McLean MA, & Sligar SG (2004). Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Letters, 556(1–3), 260–264. doi: 10.1016/s0014-5793(03)01400-5 [DOI] [PubMed] [Google Scholar]


