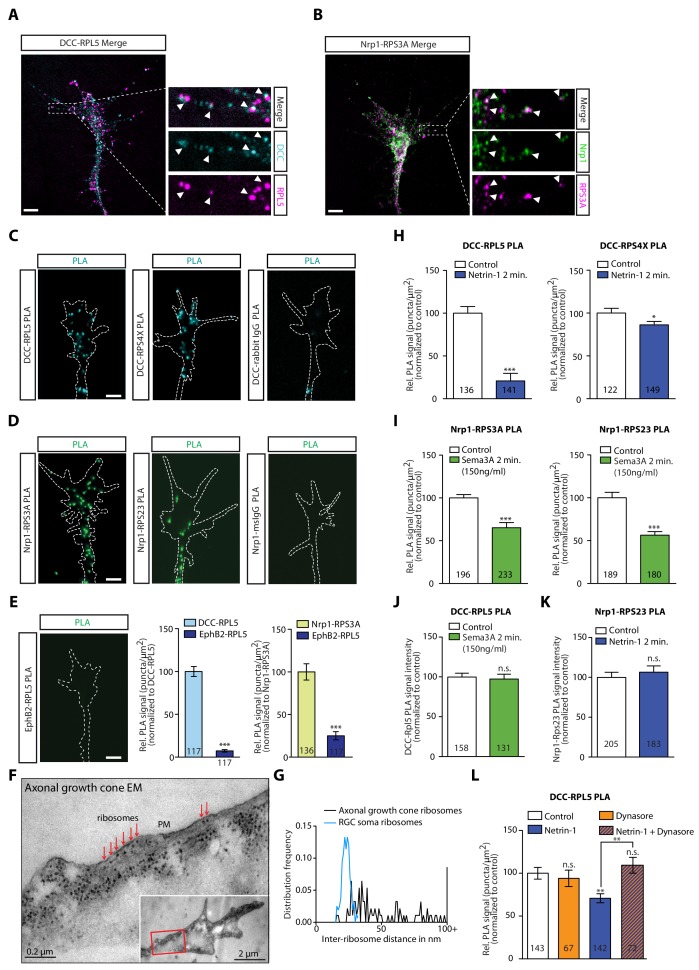
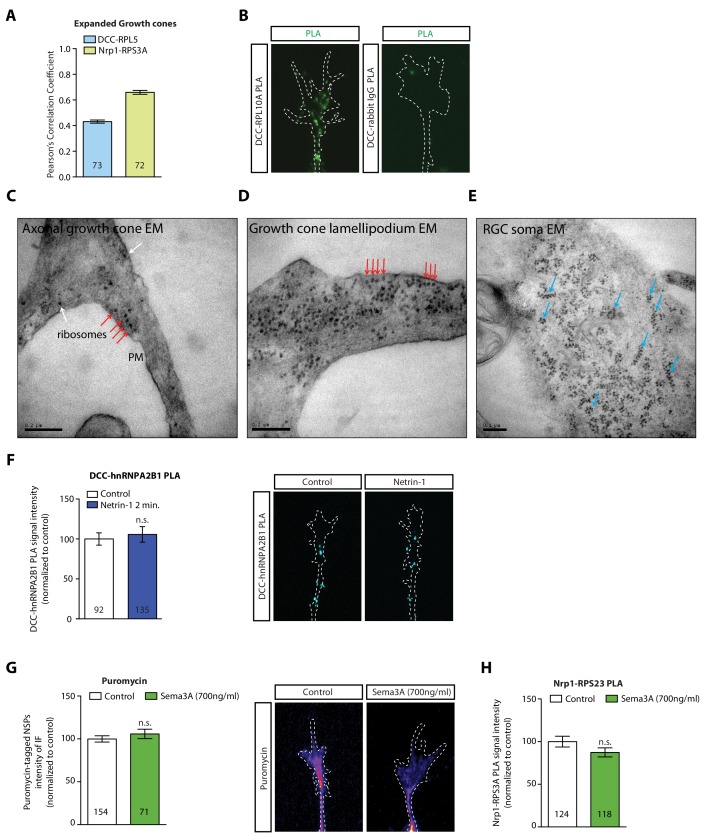
Figure 3. DCC and Nrp1 are in close proximity to ribosomes in axonal growth cones in a cue-dependent manner.
(A) Expansion imaging shows partial co-localization of DCC and (B) Nrp1 with ribosomal proteins (Scale bars, 5 μm). (C) Representative proximity ligation assay signal in axonal growth cones between DCC and RPL5/uL18, RPS4X/eS4 or IgG control (Scale bars, 5 μm). (D) Representative proximity ligation assay signal in axonal growth cones between Nrp1 and RPS3A/eS1, RPS23/uS12 or IgG control (Scale bars, 5 μm). (E) Representative PLA signal in axonal growth cones between EphB2 and RPL5/uL18 (left) and quantification of PLA signal in axonal growth cones compared to DCC-RPL5/uL18 or Nrp1-RPS23/uS12 (right) (Mann-Whitney test; three biological replicates; bars indicate mean, error bars indicate SEM, ***p<0.0001; Scale bars, 5 μm). (F) EM image of an unstimulated axonal growth cone showing ribosomes aligned in a row (red arrows) under plasma membrane (PM). Inset shows the growth cone at lower magnification; the red box indicates the area shown in higher magnification. The section glances through the extreme surface of growth cone, where it attaches to the culture dish, giving rise to areas that lack subcellular structure. (G) Distribution frequency of the inter-ribosome distance in nm of ribosomes in axonal growth cones (n = 20) or in RGC soma (n = 5). All distances larger than 100 nm were pooled together. (H, I, J, K) Quantification of PLA signal in cue-stimulated axonal growth cones relative to control (unpaired two-tailed t-test; bars indicate mean, error bars indicate SEM; ***p<0.0001; *p=0.0423; for n.s. in J p=0.3522; for n.s. in K, p=0.885). (L) Relative PLA quantification of DCC and RPL5/uL18 compared to control after Dynasore pre-treatment (50 μM for 20 min), Netrin-1, or Netrin-1 + Dynasore (one-way ANOVA with Bonferroni’s multiple comparisons test; bars indicate mean, error bars indicate SEM; p=0.001027 for Control vs. Netrin-1, p=0.000402 for Netrin-1 vs Netrin-1 + Dynasore, p=0.590377 for Control vs. Dynasore, p=0.384848 for Control vs Netrin + Dynasore). For all PLA experiments, numbers in bars indicate total number of growth cones quantified from at least three independent experiments.