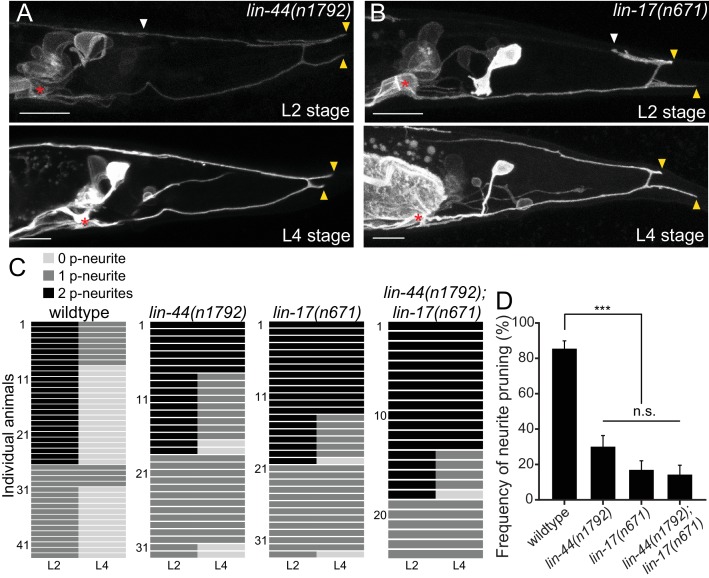
Figure 3. lin-44/wnt and lin-17/fz are required for the posterior neurite pruning.
(A and B) Representative images of single animals of lin-44(n1792) (A) and lin-17(n671) (B) mutants at L2 (top panels) and L4 (bottom panels) stages. Asterisks represent PDB cell body. White and yellow arrowheads denote anterior and posterior neurites respectively. (C) Quantification of the posterior neurite number of individual animals at L2 and L4 stages in each genetic background. Note that quantification of wildtype is from Figure 1. (D) Quantification of the posterior neurite pruning frequency. ***p<0.001; n.s., not significant (Chi-square with Yates' correction). Error bars represent standard error of proportion (SEP). Scale bars: 10 μm.