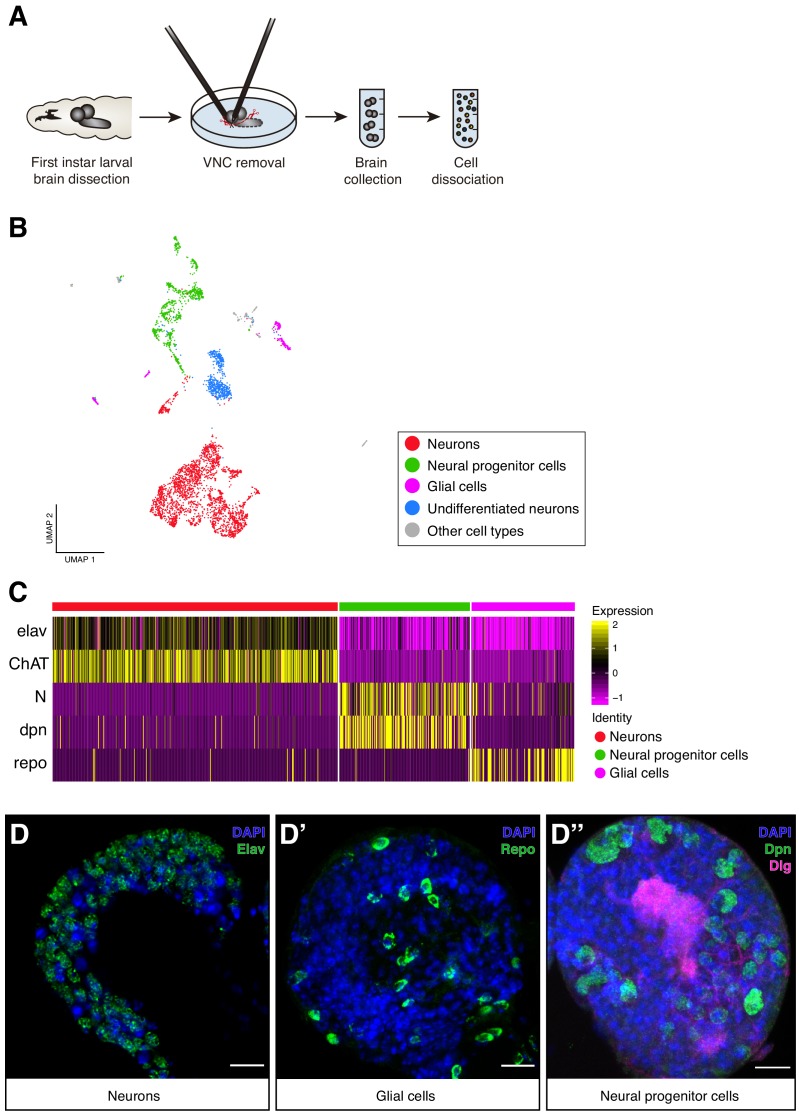
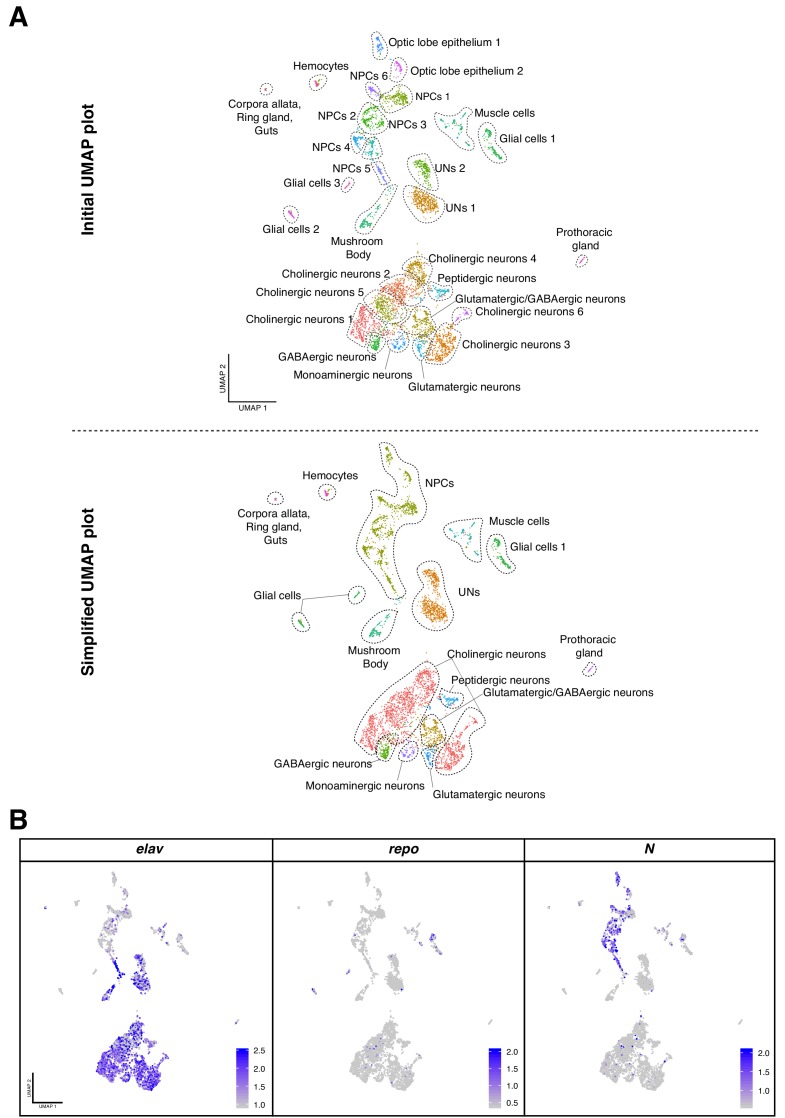
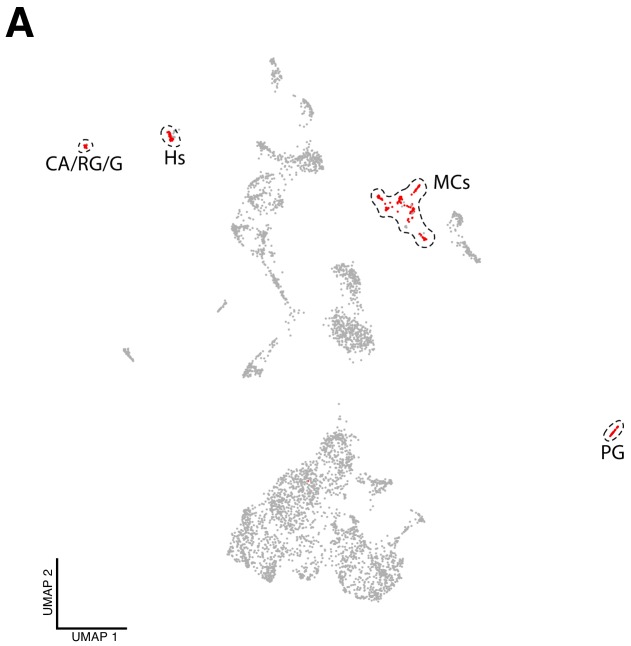
Figure 1. Cellular composition of the Larval brain by Single-Cell RNA-Seq.
(A) Experimental procedure. Drosophila late first instar larval brains/ventral nerve cords (VNC) were dissected and the VNCs were removed. Brains were collected and dissociated into a suspension of single cells. (B) Cell atlas of the larval brain reveals five main cell-types: neurons, NPCs, glial cells, UNs and other cell-types, represented in a Seurat UMAP plot. Groups are color coded. (C) Cell-types are recognized based on the expression of previously characterized marker-genes. A simplified heatmap illustrates this process: a subgroup of cholinergic neurons expresses the pan-neuronal marker elav and ChAT, a protein that catalyzes the biosynthesis of the neurotransmitter acetylcholine; NPCs are recognized by the expression of Notch and dpn and glial cells by repo expression. (D–D’’) Validation of the markers used to identify the different cell-types within the larval brain by immunostainings. The images display one lobe of the larval brain. Nuclei were labeled with DAPI and cellular borders with Dlg (disc large). Scale bar: 10 µm.