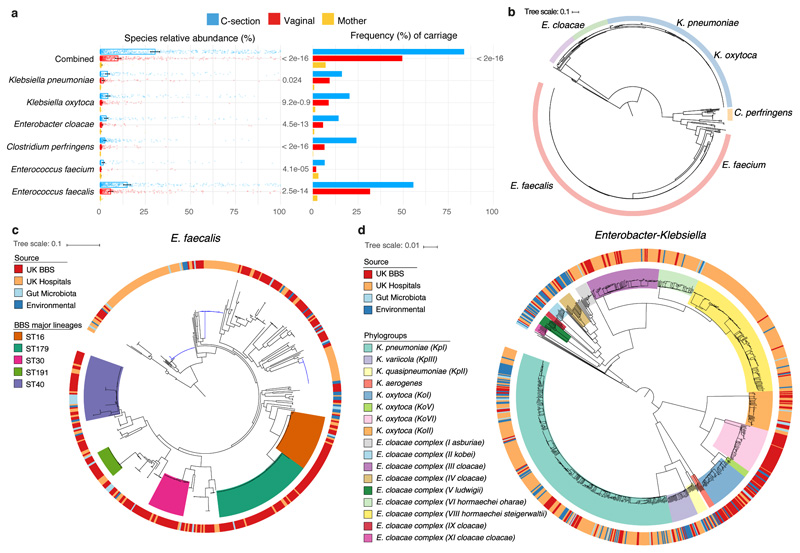
Fig. 4. Extensive and frequent colonisation of C-section delivered babies with diverse opportunistic pathogen species previously associated with healthcare infection.
a, The mean relative abundance (RA) and frequency (>1% RA) of six opportunistic pathogen species enriched in C-section born babies (n=596), compared to vaginal-born babies (n=606) during the first 21 days of life, in the context of the maternal level carriage (n=175). Error bars indicate the 95% CI of the mean RA. Statistical significance (P values indicated above) of the difference in RA and combined pathogen carriage frequency between vaginal and C-section babies was determined by two-sided Wilcoxon signed-rank test and Fisher’s exact tests, respectively. b, Phylogenetic representation of 836 bacterial strains cultured from raw faecal samples, including six opportunistic pathogens isolated five major genera: Enterococcus spp. (red, n=451); Clostridium spp. (yellow, n=24); Klebsiella spp. (blue, n=235), Enterobacter spp. (green, n=52) and Escherichia spp. (purple, n=41). c, Phylogeny of the BBS E. faecalis isolates (n=282) in the context of public isolates from the UK hospitals (n=168), human gut microbiotas (n=28) and environmental sources (n=27) with the high-risk UK epidemic lineage branches coloured in blue. Midpoint-rooted maximum likelihood tree is based on SNPs in 1,656 core genes. d, Diverse Enterobacter-Klebsiella complex strain populations among the BBS collection (n=202), in the context of the UK hospital (n=604), human gut microbiota (n=37) and environmental strains (n=120).