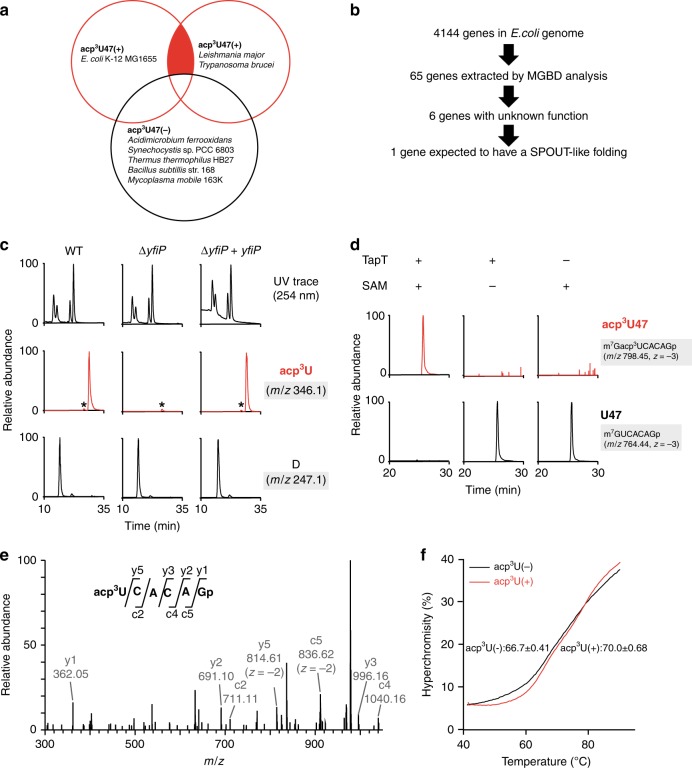
Fig. 2. yfiP is responsible for acp3U47 formation in E. coli.
a Venn diagram to narrow down the candidates by comparative genomics. Each circle represents the gene sets that are commonly present in the organisms with (+) or without (−) acp3U. Red circles represent groups of organisms that have acp3U47, whereas the black circle represents organisms that do not have acp3U47. Candidate genes lie in the red-filled region. b The process of narrowing down the candidate genes responsible for acp3U47 formation by comparative genomics. c Mass spectrometric nucleoside analyses of tRNA modifications from E. coli WT (left panels), ΔyfiP (center panels), an ΔyfiP rescued by plasmid-encoded yfiP (right panels). UV chromatogram (top) and extracted ion chromatograms (XICs) of the proton adducts of nucleosides for acp3U (middle) and D (bottom). Asterisks indicate nonspecific peaks with the same m/z value. d In vitro reconstitution of acp3U47 in E. coli tRNAMet by recombinant TapT in the presence or absence of SAM. XICs show the indicated negative ions of the RNase T1-digested fragments with (top) or without (bottom) acp3U47. e Collision-induced dissociation spectrum of the acp3U-containing fragment of the tRNAMet transcript, in which acp3U was reconstituted in vitro. The precursor ion is m/z 678.43. The product ions were assigned on the corresponding sequence as described93. The data confirmed the presence of acp3U at position 47. f Melting curves of the in vitro transcribed tRNAMet with (red lines) or without (black lines) acp3U47 modification. Tm values were determined based on the inflection point of the melting curve. Data represent average values of triplicates, with s.d. P < 4.1 × 10−3 by Student’s t test. Source data are provided as a Source Data file.