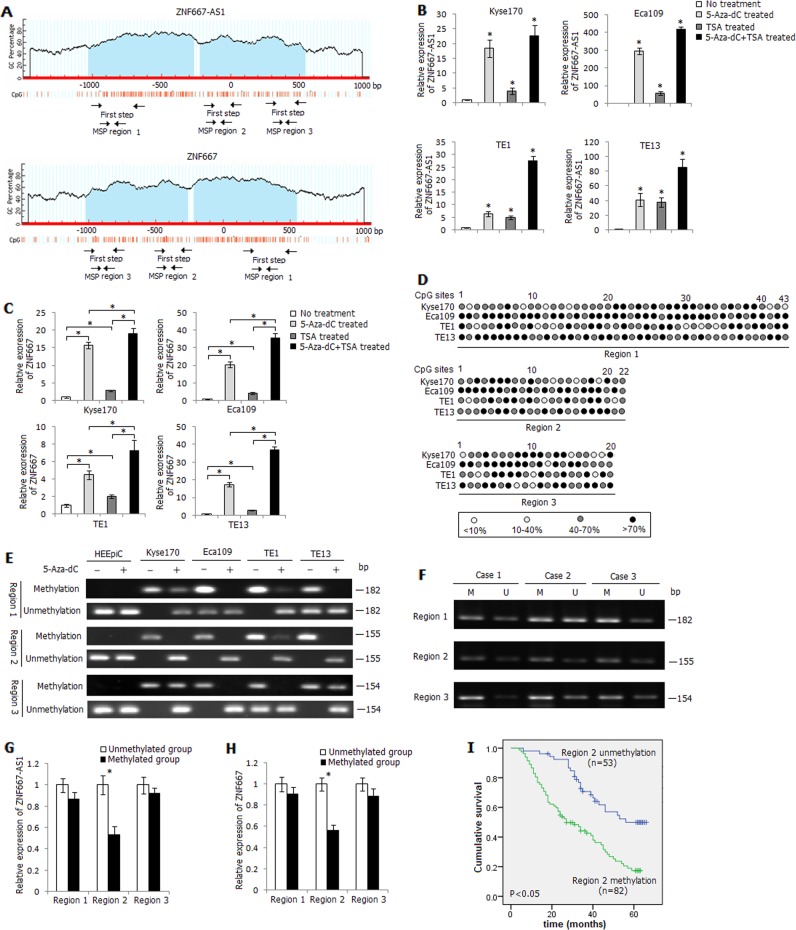
Fig. 2. Methylation status of ZNF667-AS1 and ZNF667 in ESCC.
a Schematic structure of ZNF667-AS1 and ZNF667 CpG islands predicted by MethPrimer. Three MSP regions analyzed are indicated. b Relative expression level of ZNF667-AS1 in 5-Aza-dC, TSA, or 5-Aza-dC+TSA treated esophageal cancer cells. c Relative expression level of ZNF667 in 5-Aza-dC, TSA, or 5-Aza-dC+TSA treated esophageal cancer cells. d High-resolution mapping of the methylation status of every CpG site in three regions by BGS assay in four esophageal cancer cell lines. Each CpG site is shown at the top row as an individual number. Percentage methylation was determined as percentage of methylated cytosines from 8 to 10 sequenced colonies. The color of circles for each CpG site represents the percentage of methylation. e Methylation status of three regions detected by BS-MSP assay in HEEpiC cells and esophageal cancer cells treated or untreated with 5-Aza-dC. f Methylation status of three regions detected by BS-MSP assay in ESCC tumor tissues. M: methylated; U: unmethylated. g Relative expression level of ZNF667-AS1 in tumor tissues with and without methylation of the three regions. h Relative expression level of ZNF667 in tumor tissues with and without methylation of the three regions. i The influence of region 2 methylation status on ESCC patients’ survival. *P < 0.05.