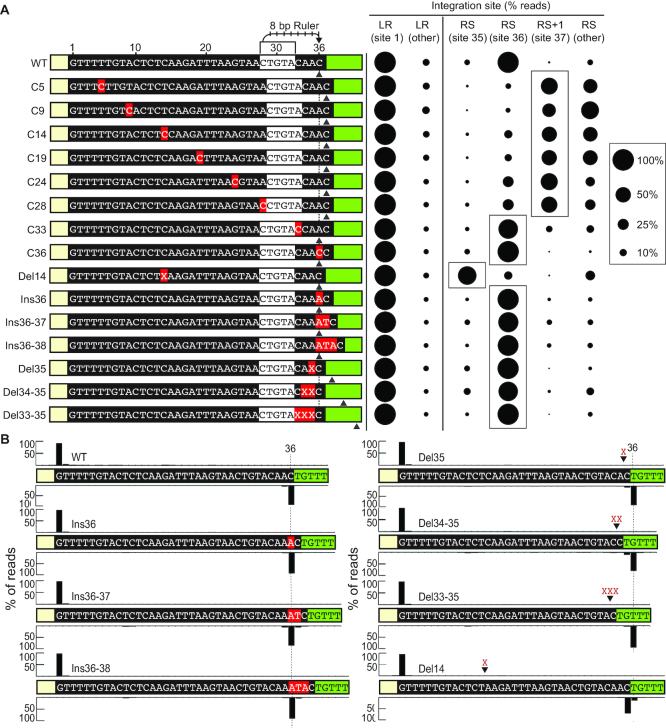
Figure 7.
Second-site integration at the repeat–spacer junction is defined by a molecular ruler. (A) (Left panel) Single, double and triple nucleotide insertion and deletion mutations made to the minimal CRISPR target. Mutations in red boxes are indicated. Predicted 5′-CTGTA-3′ ruler element defining second-site transesterification attack 8 bp from the 5′-C is marked. Dotted line marks position 36 of the repeat. Grey arrow indicates the preferred site of integration for each CRISPR target. (Right panel) Sites of integration represented as percent of total mapped reads at the leader-repeat junction (LR) and positions spanning the repeat–spacer junction (RS) qualitatively represented (right panel). Nucleotide level resolution of high-throughput sequencing data is provided in Supplemental Figure S4. (B) Integration sites for single, double and triple insertion and deletion mutations mapped to the minimal CRISPR target at nucleotide resolution. Position 36 is indicated with a dotted line. Range of total number of reads (14,884–293,723).