Figure 3.
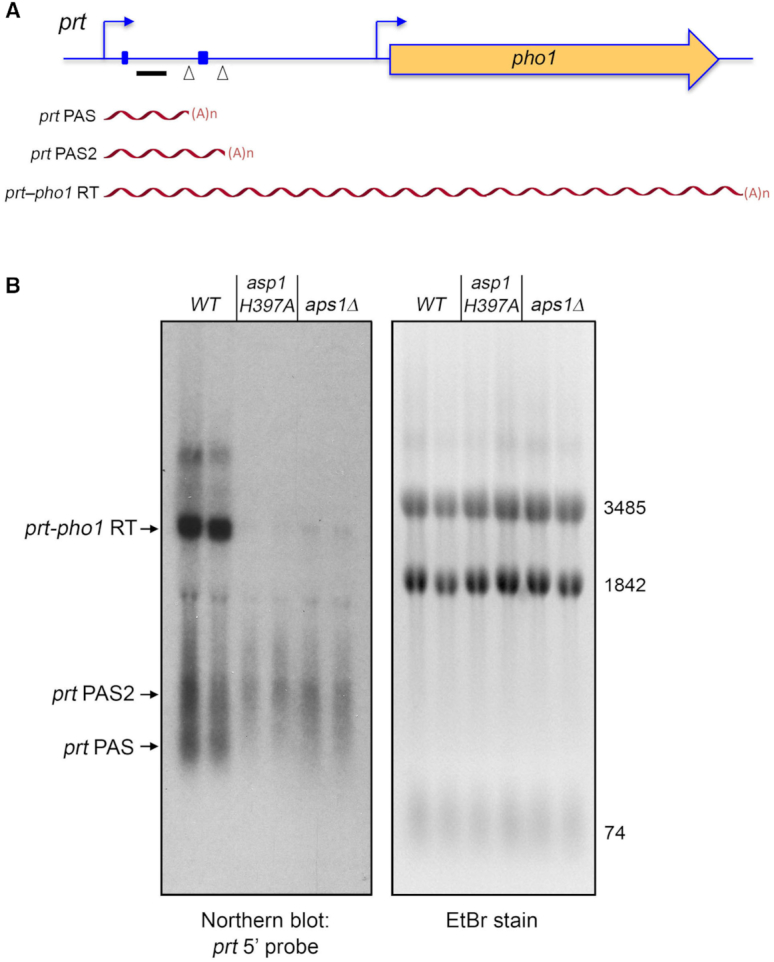
Northern analysis of prt RNA in IPP pyrophosphatase-dead mutants. (A) Schematic of the prt–pho1 locus in the reporter plasmid. Transcription start sites are indicated by bent blue arrows. Triangles denote internal poly(A) sites PAS and PAS2. DSR element clusters are indicated by small blue boxes. The gene-specific probe for prt (a 32P-labeled ssDNA complementary to the segment of the prt RNA from nucleotides +159 to +198) is denoted by a horizontal black bar. Three classes of poly(A)+prt transcripts described previously (12) are depicted as red wavy lines below the prt–pho1 locus. (B) RNA was isolated from two independent cultures of pho1Δ cells bearing the prt–pho1 reporter plasmid; the cells were either wild-type with respect to the Asp1 and Aps1 IPP pyrophosphatases or were pyrophosphatase-defective asp1-H397A or aps1Δ mutants as indicated. Right panel. The RNAs were resolved by formaldehyde-agarose gel electrophoresis and stained with ethidium bromide to visualize 28S and 18S ribosomal RNAs (3485 and 1842 nucleotides, respectively) and tRNAs (74 nucleotides). Left panel. The RNAs in the gel were transferred to membrane and hybridized to the prt probe. Annealed probe was visualized by autoradiography. The three classes of prt transcripts are indicated on the left. The PAS and PAS2 transcripts are assuredly derived from the prt–pho1 cassette, insofar as control experiments showed that they are undetectable by Northern analysis of RNA isolated from yeast strains bearing the empty plasmid vector.
