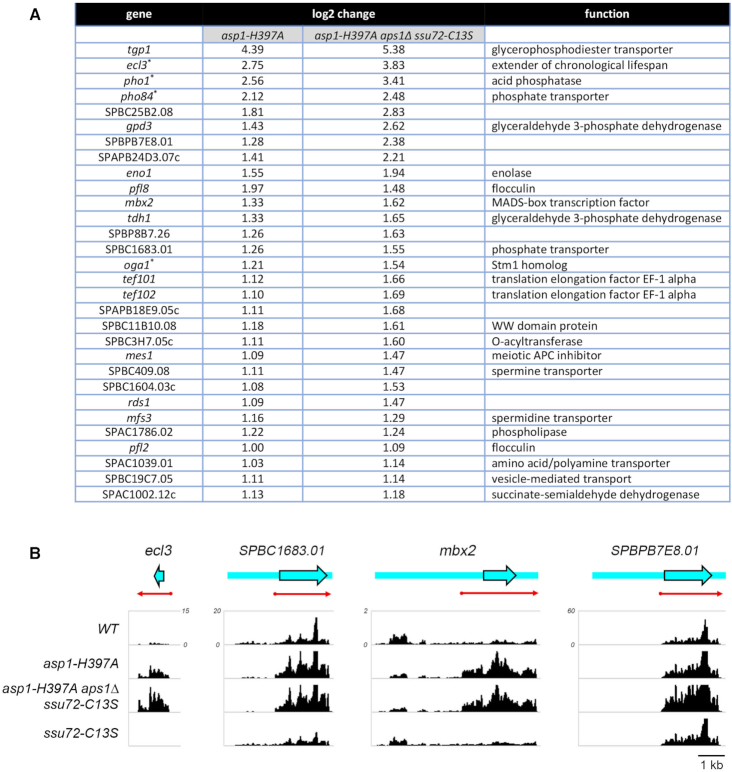
Figure 9.
RNA-seq defines an IPP-responsive gene set. (A) List of annotated protein coding genes that were up-regulated at least two-fold in asp1-H397A and asp1-H397A aps1Δ ssu72-C13S cells. Any genes that were also upregulated two-fold in ssu72-C13S cells were excluded. The four genes denoted by asterisks (pho1, pho84, ecl3, and oga1) were down-regulated by at least two-fold in the ssu72-C13S strain. (B) Strand-specific RNA-seq read densities (counts/base/million, averaged in a 25 nucleotide window) of the indicated S. pombe strains are plotted on the y-axis as a function of position across the gene loci (x-axis). The read densities were determined from cumulative counts of all three RNA-seq replicates for each S. pombe strain. The y-axis scale for all tracks of each individual gene is shown next to the WT track. The common x-axis scale is shown on the bottom right. ORFs are indicated by blue arrowheads with black outline and UTRs by blue bars extending from the ORF, shown as they are annotated in Pombase. Red arrows indicate the predicted mRNA as judged from the RNA-seq data.