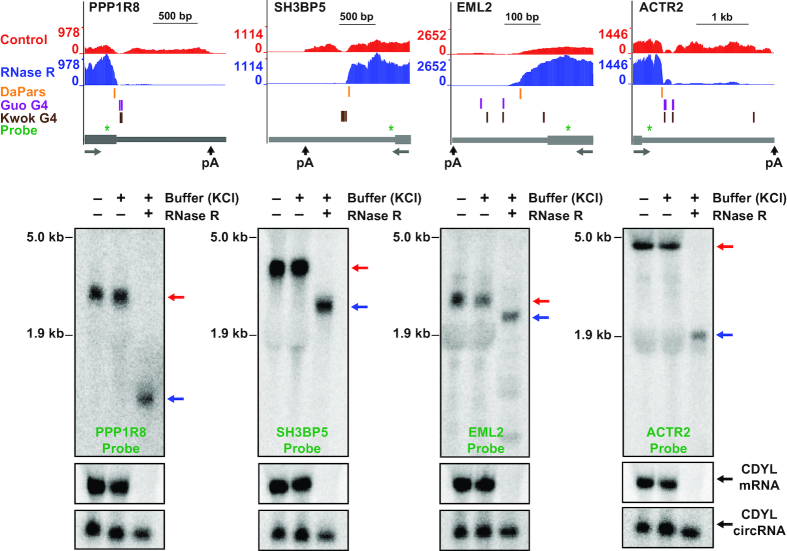
Figure 3.
RNase R stalls within the body of many mRNAs. (Top) UCSC genome browser tracks depicting the last exons of four genes that fail to be completely digested by RNase R. Gray arrows below gene models indicate the direction of transcription. RNA-seq data generated from control (red) or RNase R treated samples (blue) are shown along with RNase R stalling sites predicted by DaPars (orange), G-quadruplexes annotated by Guo and Bartel (purple) or Kwok et al. (brown), and the northern blot probes (green). (Bottom) HeLa total RNA was treated for 15 min at 37°C with either buffer (containing KCl) only or RNase R and then subjected to Northern blotting. Red and blue arrows indicate full length and partially digested transcripts, respectively. Linear and circular CDYL transcripts were used to confirm RNase R activity.