Abstract
Increasing evidence demonstrate that dysregulated microRNAs (miRNAs) are involved in carcinogenesis and tumor progression in papillary thyroid cancer (PTC). However, the specific miR-761 in cancer remains largely unknown. In this study, we reported for the first that miR-761 expression was down-regulated in PTC tissues and cell lines, and its decrease was associated with tumor size and TNM stage. Gain- and loss-of function experiments revealed that miR-761 inhibited cell proliferation, colony formation and cell cycle progression in vitro and in vivo. Moreover, TRIM29 was identified as a direct downstream target of miR-761 in PTC cells and mediated the functional effects of miR-761 in PTC. Restoration of TRIM29 expression at least partially abolished the biological effects of miR-761 on PTC cells. Furthermore, overexpression of lncRNA HOXA11-AS was inversely correlated with miR-761 expression in PTC tissues. LncRNA HOXA11-AS could modulate the miR-761 expression and regulate cellular behaviors. Taken together, this research supports the first evidence that lncRNA HOXA11-AS-reguated miR-761 plays a functional role in inhibiting PTC progression by targeting TRIM29 and represent a promising therapeutic strategy for patients with PTC.
Keywords: miR-761, thyroid cancer, TRIM29, HOXA11-AS, proliferation
Introduction
Thyroid cancer (TC) is one of the most common endocrine malignancies with a rapidly increasing incidence, which contributes to a proportion of cancer-related death worldwide, among which the papillary thyroid carcinoma (PTC) is the most common type and accounts for approximately 80% of all TC [1,2]. Despite most of the PTC are curable through thyroidectomy and conservative radioactive iodine therapy, however, tumor recurrence and distant metastasis still contribute to poor outcome [3,4]. Thus, it’s critical to understand the underlying mechanism and develop novel therapeutic strategies for its efficient treatment.
microRNAs (miRNAs), a family of highly conserved and endogenous, single stranded, small non-coding RNAs, regulates target gene expression by binding to partially complementary sequences of 3’ untranslated region (3’UTR) of target gene to induce mRNA degradation or suppresses mRNA translation [5-7]. Increasing studies confirmed that miRNAs were involved in multiple physiological and pathological activities, including cell growth, apoptosis, differentiation and metastasis [8,9]. miR-761, which located on chromosome 1p2, was dysregulated in different cancers [10-12]. miR-761 promotes progression and metastasis of non-small cell lung cancer by targeting ING4 and TIMP2 [13]. miR-761 induces aggressive phenotypes in triple-negative breast cancer cells by repressing TRIM29 expression [14]. miR-761 is up-regulated in hepatocellular carcinoma and regulates tumorigenesis by targeting mitofusin-2 [15]. These studies suggest that miR-761 was an oncogene in cancers. However, miR-761 inhibits tumor progression by targeting MSI1 in ovarian carcinoma [16]. Hsa-circ-0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells [17]. These reports indicated that miR-761 was a tumor suppressor in cancer. Above all, miR-761 was cell-type dependent in different cancers. However, the expression and its function of miR-761 in thyroid cancer remain largely unknown.
In this study, we demonstrated for the first time that miR-761 was significantly down-regulated in TC tissues and cells. Its down-regulation correlated with poor clinical features and survival. Gain- and loss-of-function experiment showed that miR-761 inhibited cell proliferation, colony formation and cell cycle progression in vitro and in vivo partly by regulating tripartite motif-containing 29 (TRIM29). miR-761 was inversely regulated by lncRNA HOXA11-AS. In conclusion, HOXA11-AS/miR-761/TRIM29 signal pathway may provide a novel and promising treatment strategy for TC.
Materials and methods
Patients and tissue samples
A total of 81 cases of PTC and corresponding adjacent normal tissues were obtained from patients who underwent thyroidectomy and node dissection, without chemotherapy or radiotherapy in our hospital. After surgical resection, the tissues were frozen in liquid nitrogen immediately and stored at -80°C until further analyses. Written consent was collected from all patients and the study was approved by the Ethics Committee of the Jilin Cancer Hospital.
Cell culture
The human thyroid cell lines, including 8505C, TPC-1, SW1736, and a human thyroid follicular epithelial cell line HTORI3 were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI1640 medium (Gibco-BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco-BRL) at 37°C in humidified atmosphere with 5% CO2.
Cell transfection and reagent
Vectors mediated precursor miR-761 (HmiR0704) and miRNA inhibitors against miR-761 (HmiR-AN1158), and their corresponding control vectors (CmiR0001-MR04 and CmiR-AN0001-AM02) were purchased from GeneCopoeia Inc. (Guangzhou, China). For TRIM29 overexpression, a full-length human TRIM29 cDNA was amplified by PCR and subcloned into the mammalian expression vector pcDNA3.1(+) (Invitrogen, Carlsbad, CA, USA). shRNA used for TRIM29 silencing and non-targeting (NT) shRNA were purchased from GenePharma (Shanghai, China). Lipofectamine 2000 Reagent (Invitrogen) was used for cell transfection. PTC cells were harvested for further analysis 48 h after transfection.
Cell proliferation and flow cytometry assay
Forty-eight hours after transfection, PTC cells (5 × 103 per well) were seeded in a 96-well plate and detected for cell proliferation using the cell counting kit-8 (CCK-8) assay according to the manufacturer’s instructions. The absorbance at 490 nm was evaluated using a Multiskan Ex microtitre plate reader (Labsystems, Helsinki, Finland) at 0 h, 24 h, 48 h, and 72 h after seeding. For cell cycle analysis, the transfected PTC cells were harvested, washed with PBS, and stained with propidium iodide (PI) using the CycleTEST PLUS DNA Reagent Kit (BD Biosciences, Franklin Lakes, NJ, USA) at room temperature for 30 min. Then the stained cells were analyzed by a FACSCalibur flow cytometer (BD Biosciences). The percentage of the cells in G1, S, and G2/M phase were analyzed.
Colony formation assay
Forty-eight hours after transfection, PTC cells (1 × 103 per well) were seeded in a 6-well plate and cultured with complete medium for 2 weeks. Cell colonies were fixed with 4% paraformaldehyde for 30 min and stained with 0.5% crystal violet for 30 min at room temperature.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA of PTC tissues and cells was isolated using Trizol reagent (Invitrogen) according to its instructions and was reverse transcribed into cDNA using a TakaRa PrimeScript™ RT kit (Ta-kara, Dalian, China). The expression of miR-761 was quantified by TaqMan miRNA assays (Applied Biosystems, Foster City, CA, USA). qRT-PCR of TRIM29 mRNA was carried out using SYBR Green Premix PCR Master Mix (Roche, Mannheim, Germany) in a StepOnePlus real-time PCR system (Applied Biosystems). The relative expression of miR-761 and TRIM29 mRNA was normalized to U6 and GAPDH using 2-ΔΔCt method.
Western blot analysis
PTC cells and tissues were lysed using RIPA lysis buffer (Beyotime, Beijing, China) on ice. An Enhanced BCA Protein Assay kit (Beyotime) was used for measuring protein concentration. The protein samples were separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). After blocking with 5% non-fat milk for 1 h at room temperature, the membranes then were incubated with specific primary antibody against TRIM29 (Abcam, Cambridge, UK) and GAPDH (sc-47724, Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 4°C overnight. The following day, the membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 2 h at room temperature. (NXA931-1ML and NA934-1ML, GE Healthcare Life Sciences, Beijing, China). The blots were detected with Chemiluminescence Reagents (Sigma, St-Louis, MO, USA) using Amersham™ Imager 680 from GE Healthcare Life Sciences.
Tumor xenograft model
BALB/c nude mice (male, 4-6-week-old, 18-20 g) were obtained from Shanghai SLAC Laboratory Animal Co. Ltd (Shanghai, China) and randomly divided into two groups (n = 5 per group). TPC-1 cells (1 × 106 per injection) that were transfected with control and miR-761 were implanted into the right flank of the mice via subcutaneous injection. Tumor volumes were measured every 4 days after being apparently observed and calculated with the following formula: Volume = (length × width2)/2. After 3 weeks, all mice were sacrificed under anesthesia. The animal experiments were approved by the Animal Care and Use Committee of Jilin Cancer Hospital.
Luciferase reporter assay
The 3’-UTR of TRIM29 containing predicted miR-761 binding sites (both wild type and mutant) were subcloned into the pmirGLO vector (Promega, Madison, WI, USA). For the luciferase reporter assay, TPC-1 cells were transfected with different combinations of miR-761 mimics, control mimics and pGL3-TRIM29 3’-UTR wild type or mutant. The relative luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) and normalized to Renilla activity.
Statistical analysis
The data were shown as Mean ± SD and analyzed using GraphPad Prism 6.0 Software (GraphPad Inc., San Diego, CA, USA). All experiments were repeated for at least 3 times. Student’s t-test (2 groups) and one-way ANOVA (multiple groups) were conducted to analyze the difference. Spearman Pearson correlation analysis was performed to determine the correlation between miR-761 and TRIM29 mRNA expression. Differences were defined as statistically significant if P < 0.05.
Results
miR-761 was down-regulated in PTC and correlated with poor clinicopathological features
To examine the expression of miR-761 in PTC, we demonstrated that miR-761 was significantly decreased in PTC tissues compared to that in adjacent non-tumor tissues using qRT-PCR (P < 0.05, Figure 1A). Moreover, miR-761 was also down-regulated in a panel of PTC cell lines compared with normal thyroid cell HTORI3 (P < 0.05, Figure 1B). To assess the clinical roles of miR-761, PTC tissues were divided into two subgroups according to the median value. The decreased miR-761 was significantly associated with large tumor size and advanced TNM stage (P = 0.008, 0.038, respectively, Table 1). These results suggested that miR-761 was involved in PTC progression.
Figure 1.
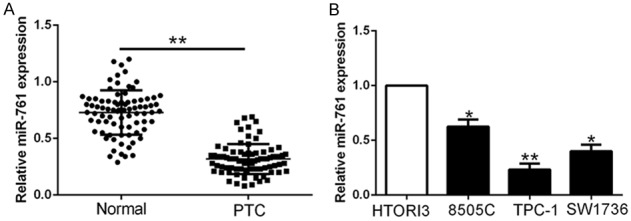
Expression of miR-761 in PTC. A. The expression level of miR-761 was assessed in tumor tissues and adjacent normal tissues of PTC patients. B. The expression levels of miR-761 were down-regulated in PTC cells lines compared to normal thyroid cell line HTORI3. *P < 0.05, **P < 0.01.
Table 1.
Correlation between miR-761 expression and clinicopathological feature in PTC patients (n = 81)
| Clinical parameters | Cases (n) | Expression level | P value | |
|---|---|---|---|---|
|
| ||||
| miR-761high (n = 40) | miR-761low (n = 41) | |||
| Age (years) | ||||
| < 60 years | 32 | 17 | 15 | 0.586 |
| ≥ 60 years | 49 | 23 | 26 | |
| Gender | ||||
| Male | 36 | 18 | 18 | 0.921 |
| Female | 45 | 22 | 23 | |
| Tumor size (cm) | 0.008* | |||
| < 5 cm | 58 | 34 | 24 | |
| ≥ 5 cm | 23 | 6 | 17 | |
| TNM stage | 0.038* | |||
| I+II | 63 | 35 | 28 | |
| III+IV | 18 | 5 | 13 | |
| Lymph metastasis | 0.615 | |||
| Present | 16 | 7 | 9 | |
| Absent | 65 | 33 | 32 | |
indicate P < 0.05.
miR-761 inhibits cell proliferation, colony formation and cell cycle progression in thyroid cancer
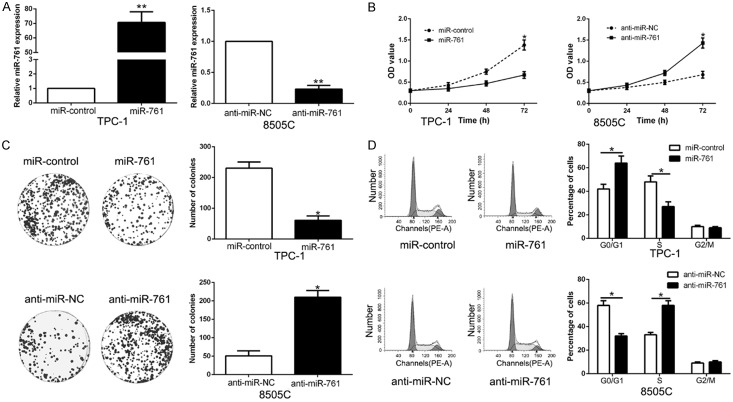
To determine the function of miR-761 in PTC, we performed gain- and loss-of function experiment by transfecting respective vectors in TPC-1 and 8505C whose endogenous miR-761 was lowest and highest expression in PTC cell lines (P < 0.05, Figure 2A). The results of CCK-8, colony formation and flow cytometry assay showed that miR-761 overexpression significantly repressed the cell proliferation, colony formation and cell cycle progression in TPC-1 cells (P < 0.05, Figure 2B-D). While miR-761 knockdown had the opposite effect on 8505C cells (P < 0.05, Figure 2B-D). These data suggest that miR-761 exhibits a tumor suppressor role in PTC.
Figure 2.
miR-761 suppresses proliferation, colony formation and cell cycle progression of PTC cells. (A) TPC-1 and 8505C cells that were transfected with different vectors were subjected to qRT-PCR for miR-761 expression. Overexpression of miR-761 inhibited cell proliferation (B), colony formation (C) and cell cycle progression (D) in TPC-1 cells, while down-regulation of miR-761 promoted proliferation (B), cell colony formation (C) and cell cycle progression (D) in 8505C cells. n = six independent experiments. *P < 0.05, **P < 0.01.
miR-761 overexpression restrains PTC growth in vivo
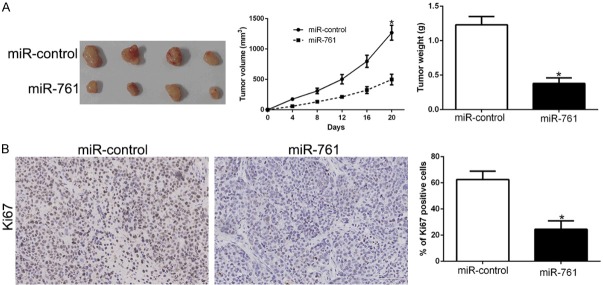
Subcutaneous tumor model was conducted to determine the effects of miR-761 in vivo. The tumors of xenograft mice models were shown in Figure 3A. Consistent with in vitro data, the tumor growth curves showed that miR-761 overexpression inhibited PTC growth measuring by tumor size and weight in mice (P < 0.05, Figure 3A). Moreover, immunohistochemistry for Ki-67 revealed that miR-761 decrease the proportion of Ki-67 positive cells (P < 0.05, Figure 3B). These data indicate that miR-761 contributes to PTC tumorigenesis in vivo.
Figure 3.
miR-761 inhibits tumor growth in vivo. A. Tumor growth curve revealed that miR-761 overexpression significantly inhibited tumor growth in vivo. B. Tumor nodules were subjected to immunohistochemical staining for Ki-67 assays and quantitative analysis. *P < 0.05.
TRIM29 was a direct target of miR-761 in PTC cells
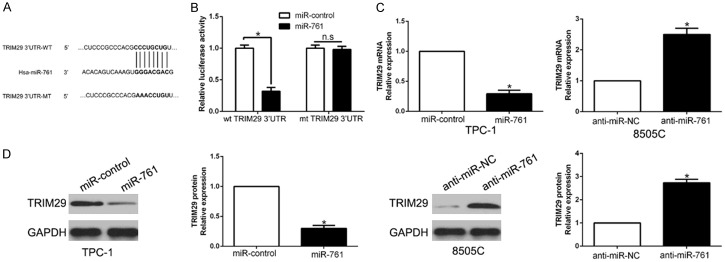
To investigate the mechanism of miR-761 in PTC, we used algorithm TargetScan to search potential target of miR-761 and the TRIM29 3’-UTR had the binding site of miR-761 (Figure 4A). To confirm whether TRIM29 was a target of miR-761 in PTC cells, we performed luciferase reporter assay to show that miR-761 overexpression obviously inhibited the luciferase activity of wild-type (wt) TRIM29 3’-UTR while had no influence on that of mutant (mt) TRIM29 3’-UTR (P < 0.05, Figure 4B). Furthermore, miR-761 overexpression markedly suppressed the mRNA and protein expression of TRIM29 in TPC-1 cells (P < 0.05, respectively, Figure 4C, 4D). While the expression of TRIM29 mRNA and protein were significantly increased by the inhibition of miR-761 in 8505C cells (P < 0.05, respectively, Figure 4C, 4D). Moreover, we confirmed that TRIM29 was up-regulated in PTC tissues compared to the normal tissues (Figure S1B).
Figure 4.
TRIM29 is a direct target of miR-761 in PTC cells. (A) miR-761 and its putative binding sequences in the 3’-UTR of TRIM29. The mutant binding site was generated in the complementary site for the seed region of miR-761. (B) miR-761 overexpression significantly suppressed the luciferase activity that carried wild-type (wt) but not mutant (mt) 3’-UTR of TRIM29. miR-761 overexpression reduced the expression of TRIM29 mRNA (C) and protein (D) in TPC-1 cells and miR-761 knockdown increased the level of TRIM29 mRNA (C) and protein (D) in 8505C cells. *P < 0.05.
miR-761 expression was inversely correlated with TRIM29 in PTC tissues
To further validate the correlation between miR-761 and TRIM29 in vivo, we examined the TRIM29 mRNA and protein expression in two miR-761 expression subgroups. Results showed that TRIM29 mRNA and protein levels were significantly lower in high miR-761 group than that in low miR-761 group in PTC (P < 0.05, Figure 5A, 5B). In addition, we demonstrated that the mRNA level of TRIM29 in the PTC tissues was inversely correlated with miR-761 expression (r = -0.6207, P < 0.0001, Figure 5C). Moreover, we used IHC staining of TRIM29 in different miR-761 tissues, which showed the similar result (Figure S1A). In conclusion, these data suggest that TRIM29 was a direct downstream target of miR-761 in PTC.
Figure 5.
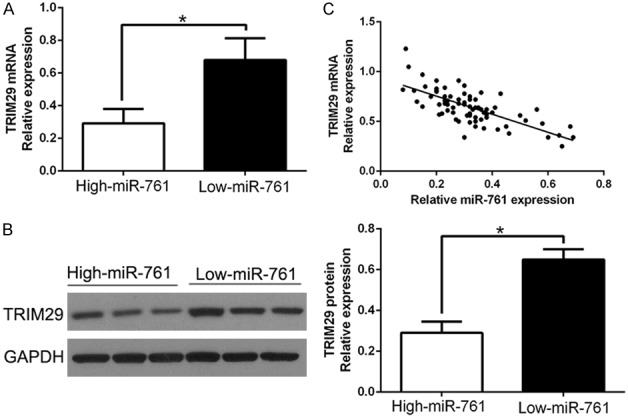
An inverse correlation between miR-761 and TRIM29 expression is observed in PTC. A. The expression of TRIM29 mRNA in miR-761 high-expressing tumors was significantly lower than that in miR-761 low-expressing tumors. B. The expression of TRIM29 protein in miR-761 high-expressing tumors was significantly lower than that in miR-761 low-expressing tumors. C. A significant inverse correlation between the mRNA levels of TRIM29 and miR-761 was observed in PTC tissues. *P < 0.05.
TRIM29 promotes cell proliferation, colony formation and cell cycle progression in thyroid cancer
To explore whether TRIM29 overexpression mimic the biological function of miR-761, we used overexpressed or knockdown vectors to mimic the experiment (P < 0.05, Figure 6A). The data showed that TRIM29 overexpression significantly promoted the cell proliferation, colony formation and cell cycle progression in 8505C cells (P < 0.05, Figure 6B-D). While TRIM29 knockdown had the opposite effect on TPC-1 cells (P < 0.05, Figure 6B-D). Thus, these data indicated that TRIM29 alteration could mimic the biological function of miR-761 in PTC cells.
Figure 6.
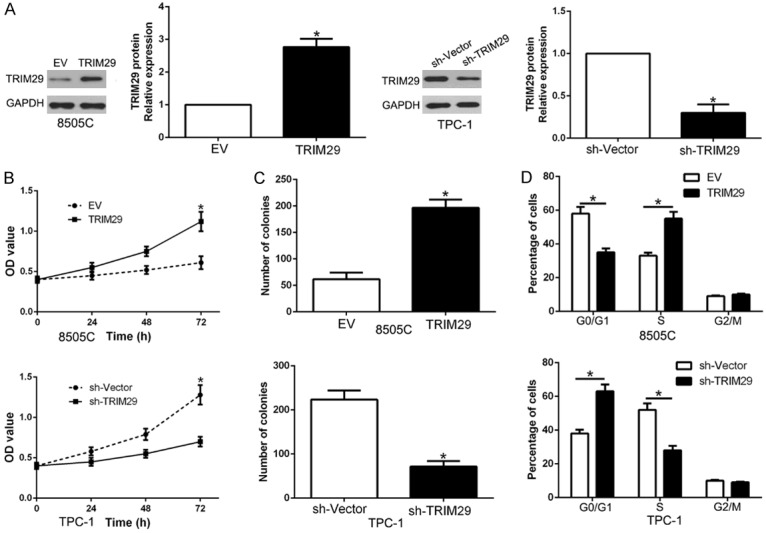
TRIM29 promotes cell proliferation, colony formation and cell cycle progression in PTC cells in vitro. (A) 8505C and TPC1 cells that were transfected with corresponding vectors were subjected to western blot for TRIM29 expression. Overexpression of TRIM29 promoted cell proliferation (B), colony formation (C) and cell cycle progression (D) in 8505C cells, while down-regulation of TRIM29 inhibited proliferation (B), cell colony formation (C) and cell cycle progression in TPC-1 cells. n = six independent experiments. *P < 0.05.
TRIM29 mediates the functional role of miR-761 in PTC cells
To detect whether TRIM29 participates in the effects of miR-761 on PTC cells, TRIM29 was overexpressed in TPC-1-miR-761 cells or knock down TRIM29 in 8505C-anti-miR-761 cells (P < 0.05, Figure 7A). TRIM29 restoration significantly abolished the inhibitory effects of miR-761 on cell proliferation, colony formation and cell cycle progression of TPC-1 cells (P < 0.05, Figure 7B-D). Moreover, the effects of miR-761 knockdown on cell proliferation, colony formation and cell cycle progression were attenuated by co-transfection with TRIM29 shRNA (P < 0.05, Figure 7B-D). These results indicate that TRIM29 is essential for miR-761-induced functional effects on PTC cells.
Figure 7.
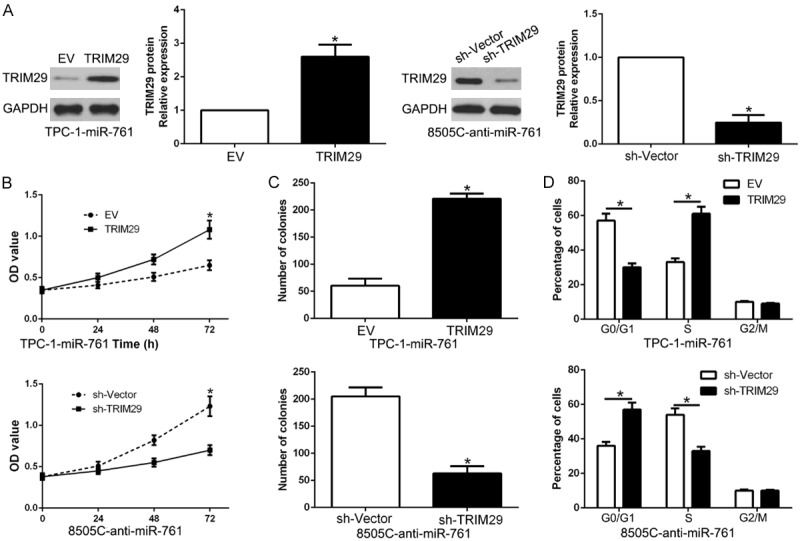
Modulation of TRIM29 partially abolishes miR-761 mediated cellular processes in PTC. (A) miR-761-overexpressing TPC-1 cells that were transfected with empty vector (EV) or HMGA2 overexpression vector were subjected to western blot for TRIM29. miR-761-suppressive 8505C cells that were transfected with control shRNA or TRIM29 shRNA were subjected to western blot for TRIM29. TRIM29 restoration abrogated the effects of miR-761 overexpression on proliferation (B), cell colony formation (C) and cell cycle progression (D) of TPC-1 cells. TRIM29 knockdown reversed the promotive effects of miR-761 knockdown in 8505C cells (B-D). *P < 0.05.
miR-761 is negatively regulated by lncRNA HOXA11-AS in PTC cells
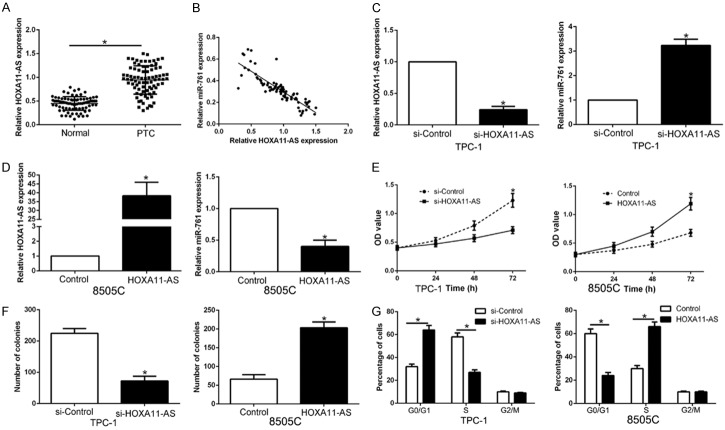
To investigate the mechanism of miR-761 was down-regulated in PTC, we predicted a target by Starbase v2.0 and found lncRNA HOXA11-AS is a molecular sponge which may regulate miR-761. We first determined qRT-PCR and found that HOXA11-AS was significantly increased in PTC tissues compared to those adjacent non-tumor tissues (P < 0.05, Figure 8A). Spearman analysis revealed that HOXA11-AS was inversely correlated with miR-761 in PTC tissues (r = -0.7132, P < 0.05, Figure 8B). Furthermore, HXOA11-AS knockdown increased the expression of miR-761 in TPC-1 cells (P < 0.05, Figure 8C), While HOXA11-AS overexpression led a reduction of miR-761 in 8505C cells (P < 0.05, Figure 8D). Then, we investigated whether HOXA11-AS regulated biological function in PTC cells. As expect, HOXA11-AS knockdown inhibits cell proliferation, colony formation and cell cycle progression in TPC-1 cells (P < 0.05, Figure 8E-G). On the contrary, HOXA11-AS overexpression promoted cell proliferation, colony formation and cell cycle progression in 8505C cells (P < 0.05, Figure 8E-G). Therefore, lncRNA HOXA11-AS exerts its function in PTC cells possibly by negatively regulating miR-761.
Figure 8.
miR-761 is negatively regulated by lncRNA HOXA11-AS in PTC cells. (A) LncRNA HOXA11-AS expression differences between PTC tissues and adjacent non-tumor tissues. (B) An inverse correlation between the levels of miR-761 and HOXA11-AS expression was observed in PTC tissues. (C) TPC-1 that were transfected with HOXA11-AS siRNA or control siRNA were detected by qRT-PCR. HOXA11-AS knockdown significantly increased the expression of miR-761 in TPC-1 cells. (D) 8505C cells that were transfected with HOXA11-AS vector or empty vector were confirmed by qRT-PCR. HOXA11-AS overexpression notably reduced the expression of miR-761 in 8505C cells. Down-regulation of HOXA11-AS inhibited cell proliferation (E), colon formation (F) and cell cycle progression (G) in TPC-1 cells, while overexpression of HOXA11-AS promoted cell proliferation (E), colon formation (F) and cell cycle progression (G) in 8505C cells. n = 6 independent experiments. *P < 0.05.
Discussion
Increasing evidence reported that miRNAs play critical role in the initiation and progression of thyroid cancer by regulating target oncogene or tumor suppressor which are involved in cancer pathogenesis. Therefore, identification of cancer-specific miRNAs may contribute to provide novel diagnosis biomarkers and therapeutic targets [18]. MiR-761, which was a novel cancer-related miRNA, the expression and functional importance in TC remains unknown. Here, we demonstrated that miR-761 was significantly decreased in PTC tissues and cell lines, which correlated with poor clinical features of patients. Moreover, functional experiments confirmed that miR-761 inhibited proliferation, colony formation and cell cycle progression by gain- and loss-of-function experiment. In addition, we also confirmed that miR-761 restrained tumor growth in vivo. These results revealed that miR-761 act as a tumor suppressor in PTC progression.
TRIM29, which has been recognized as a novel oncogene in PTC [19]. TRIM29 promotes progression of thyroid carcinoma via activating PI3K/AKT signaling pathway [19]. Previous studies also reported that TRIM29 was involved different cancers [20-22]. TRIM29 overexpression promotes proliferation and survival of bladder cancer cells through NF-κB signaling [23]. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells [24]. Here, we demonstrated that miR-761 repressed TRIM29 expression via binding its 3’UTR. In PTC tissues, miR-761 was inversely correlated with TRIM29 expression. TRIM29 also promoted PTC cell proliferation, colony formation and cell cycle progression. Moreover, TRIM29 restoration reversed the effects of miR-761 on PTC cells. Interestingly, TRIM29 was post-transcriptionally regulated by miR-761 breast cancer [14]. These data suggest that miR-761/TRIM29 axis exert important roles in PTC.
To understand the reason that miR-761 was down-regulated in PTC, we searched public database and showed that lncRNA HOXA11-AS maybe a sponge of miR-761. HOXA11-AS has been identified as an oncogene in cancers by regulating miRNA [25-28]. LncRNA HOXA11-AS promotes proliferation and invasion by targeting miR-124 in human non-small cell lung cancer cells [29]. LncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer [30]. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p [31]. Here, we confirmed that HOXA11-AS was up-regulated in PTC and negatively correlated with miR-761 expression. HOXA11-AS could negatively regulated miR-761 expression in PTC cells and exerted its function in proliferation, colony formation and cell cycle progression. These data suggest that HOXA11-AS play an oncogene role in PTC through regulating miR-761.
To conclude, our results demonstrated that miR-761 expression was down-regulated in PTC for the first time. Furthermore, miR-761 inhibited PTC cell proliferation, colony formation and cell cycle progression through binding to the 3’UTR of TRIM29 and subsequently repressing its expression. Finally, we confirmed that HOXA11-AS could regulated miR-761 expression. Thus, our results imply that miR-761 functions as a tumor suppressor in PTC and may be a promising therapeutic target for PTC treatment.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Fagin JA, Wells SA Jr. Biologic and clinical perspectives on thyroid cancer. N Engl J Med. 2016;375:1054–1067. doi: 10.1056/NEJMra1501993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaijee F, Nikiforov YE. Molecular analysis of thyroid tumors. Endocr Pathol. 2011;22:126–133. doi: 10.1007/s12022-011-9170-y. [DOI] [PubMed] [Google Scholar]
- 3.Sherman SI. Thyroid carcinoma. Lancet. 2003;361:501–511. doi: 10.1016/s0140-6736(03)12488-9. [DOI] [PubMed] [Google Scholar]
- 4.Pellegriti G, Frasca F, Regalbuto C, Squatrito S, Vigneri R. Worldwide increasing incidence of thyroid cancer: update on epidemiology and risk factors. J Cancer Epidemiol. 2013;2013:965212. doi: 10.1155/2013/965212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Yuan JH, Huang JF, Yang F, Wang TT, Ma JZ, Zhang L, Zhou CC, Wang F, Yu J, Zhou WP, Sun SH. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Dou C, Yao B, Xu M, Ding L, Wang Y, Jia Y, Li Q, Zhang H, Tu K, Song T, Liu Q. Methylation-mediated repression of microRNA-129-2 suppresses cell aggressiveness by inhibiting high mobility group box 1 in human hepatocellular carcinoma. Oncotarget. 2016;7:36909–36923. doi: 10.18632/oncotarget.9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiozawa K, Shuting J, Yoshioka Y, Ochiya T, Kondo T. Extracellular vesicle-encapsulated microRNA-761 enhances pazopanib resistance in synovial sarcoma. Biochem Biophys Res Commun. 2018;495:1322–1327. doi: 10.1016/j.bbrc.2017.11.164. [DOI] [PubMed] [Google Scholar]
- 11.Cao S, Lin L, Xia X, Wu H. MicroRNA-761 promotes the sensitivity of colorectal cancer cells to 5-Fluorouracil through targeting FOXM1. Oncotarget. 2018;9:321–331. doi: 10.18632/oncotarget.20109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long B, Wang K, Li N, Murtaza I, Xiao JY, Fan YY, Liu CY, Li WH, Cheng Z, Li P. miR-761 regulates the mitochondrial network by targeting mitochondrial fission factor. Free Radic Biol Med. 2013;65:371–379. doi: 10.1016/j.freeradbiomed.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan A, Yang C, Chen Z, Li C, Cai L. MiR-761 promotes progression and metastasis of non-small cell lung cancer by targeting ING4 and TIMP2. Cell Physiol Biochem. 2015;37:55–66. doi: 10.1159/000430333. [DOI] [PubMed] [Google Scholar]
- 14.Guo GC, Wang JX, Han ML, Zhang LP, Li L. microRNA-761 induces aggressive phenotypes in triple-negative breast cancer cells by repressing TRIM29 expression. Cell Oncol (Dordr) 2017;40:157–166. doi: 10.1007/s13402-016-0312-6. [DOI] [PubMed] [Google Scholar]
- 15.Zhou X, Zhang L, Zheng B, Yan Y, Zhang Y, Xie H, Zhou L, Zheng S, Wang W. MicroRNA-761 is upregulated in hepatocellular carcinoma and regulates tumorigenesis by targeting Mitofusin-2. Cancer Sci. 2016;107:424–432. doi: 10.1111/cas.12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi C, Zhang Z. miR-761 inhibits tumor progression by targeting MSI1 in ovarian carcinoma. Tumour Biol. 2016;37:5437–5443. doi: 10.1007/s13277-015-4377-z. [DOI] [PubMed] [Google Scholar]
- 17.Li GF, Li L, Yao ZQ, Zhuang SJ. Hsa_circ_0007534/miR-761/ZIC5 regulatory loop modulates the proliferation and migration of glioma cells. Biochem Biophys Res Commun. 2018;499:765–771. doi: 10.1016/j.bbrc.2018.03.219. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Wang Y, Dou C, Sun L, Li Q, Wang L, Xu Q, Yang W, Liu Q, Tu K. MicroRNA-1468 promotes tumor progression by activating PPAR-gamma-mediated AKT signaling in human hepatocellular carcinoma. J Exp Clin Cancer Res. 2018;37:49. doi: 10.1186/s13046-018-0717-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Xu J, Li Z, Su Q, Zhao J, Ma J. TRIM29 promotes progression of thyroid carcinoma via activating P13K/AKT signaling pathway. Oncol Rep. 2017;37:1555–1564. doi: 10.3892/or.2017.5364. [DOI] [PubMed] [Google Scholar]
- 20.Liang C, Dong H, Miao C, Zhu J, Wang J, Li P, Li J, Wang Z. TRIM29 as a prognostic predictor for multiple human malignant neoplasms: a systematic review and meta-analysis. Oncotarget. 2018;9:12323–12332. doi: 10.18632/oncotarget.23617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng SX, Cai QC, Guo CH, Zhi LQ, Dai X, Zhang DF, Ma W. High expression of TRIM29 (ATDC) contributes to poor prognosis and tumor metastasis by inducing epithelialmesenchymal transition in osteosarcoma. Oncol Rep. 2017;38:1645–1654. doi: 10.3892/or.2017.5842. [DOI] [PubMed] [Google Scholar]
- 22.Dukel M, Streitfeld WS, Tang TC, Backman LR, Ai L, May WS, Brown KD. The breast cancer tumor suppressor TRIM29 is expressed via ATM-dependent signaling in response to hypoxia. J Biol Chem. 2016;291:21541–21552. doi: 10.1074/jbc.M116.730960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan ST, Liu SY, Wu B. TRIM29 overexpression promotes proliferation and survival of bladder cancer cells through NF-kappaB signaling. Cancer Res Treat. 2016;48:1302–1312. doi: 10.4143/crt.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu W, Xu B, Yao Y, Yu X, Cao H, Zhang J, Liu J, Sheng H. RNA interference against TRIM29 inhibits migration and invasion of colorectal cancer cells. Oncol Rep. 2016;36:1411–1418. doi: 10.3892/or.2016.4941. [DOI] [PubMed] [Google Scholar]
- 25.Qu L, Jin M, Yang L, Sun C, Wang P, Li Y, Tian L, Liu M, Sun Y. Expression of long non-coding RNA HOXA11-AS is correlated with progression of laryngeal squamous cell carcinoma. Am J Transl Res. 2018;10:573–580. [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Y, Yi L, Zhao JZ, Jiang YG. Long noncoding RNA HOXA11-AS functions as miRNA sponge to promote the glioma tumorigenesis through targeting miR-140-5p. DNA Cell Biol. 2017;36:822–828. doi: 10.1089/dna.2017.3805. [DOI] [PubMed] [Google Scholar]
- 27.Li W, Jia G, Qu Y, Du Q, Liu B, Liu B. Long Non-Coding RNA (LncRNA) HOXA11-AS promotes breast cancer invasion and metastasis by regulating epithelial-mesenchymal transition. Med Sci Monit. 2017;23:3393–3403. doi: 10.12659/MSM.904892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Z, Chen Z, Fan R, Jiang B, Chen X, Chen Q, Nie F, Lu K, Sun M. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer. 2017;16:82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu W, Peng W, Jiang H, Sha H, Li J. LncRNA HOXA11-AS promotes proliferation and invasion by targeting miR-124 in human non-small cell lung cancer cells. Tumour Biol. 2017;39:1010428317721440. doi: 10.1177/1010428317721440. [DOI] [PubMed] [Google Scholar]
- 30.Chen D, Sun Q, Zhang L, Zhou X, Cheng X, Zhou D, Ye F, Lin J, Wang W. The lncRNA HOXA11-AS functions as a competing endogenous RNA to regulate PADI2 expression by sponging miR-125a-5p in liver metastasis of colorectal cancer. Oncotarget. 2017;8:70642–70652. doi: 10.18632/oncotarget.19956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhan M, He K, Xiao J, Liu F, Wang H, Xia Z, Duan X, Huang R, Li Y, He X, Yin H, Xiang G, Lu L. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018 doi: 10.1111/jcmm.13633. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.