-
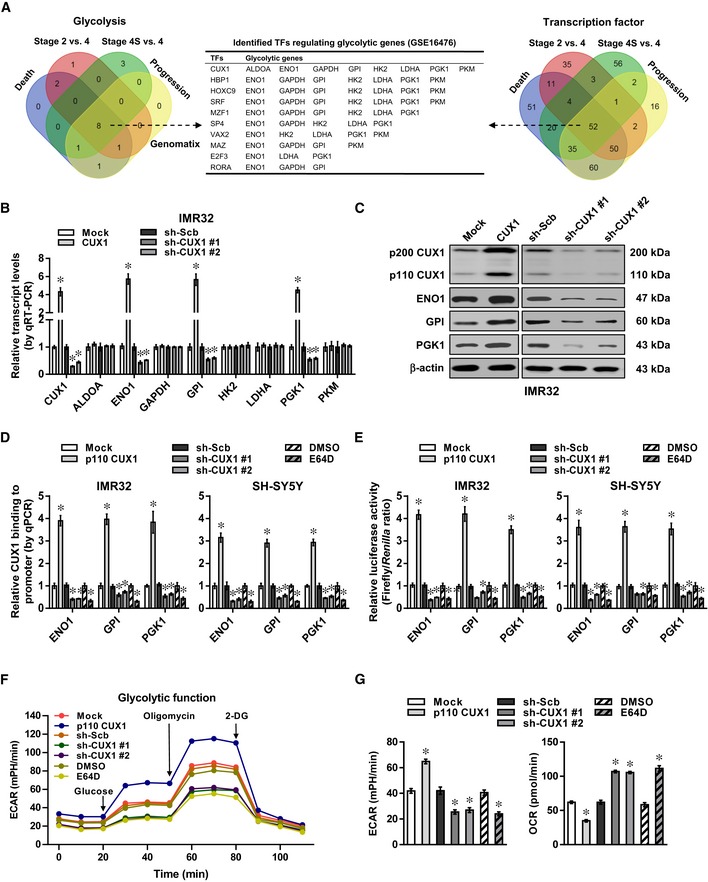
A
Venn diagram indicating the identification of differentially expressed glycolytic genes and transcription factors (fold change > 2.0, Student's
t‐test,
P < 0.05) in 88 NB cases (
GSE16476), and over‐lapping analysis with potential transcription factors regulating glycolytic genes revealed by Genomatix program.
-
B, C
Real‐time qRT–PCR (B, normalized to β‐actin, n = 5) and Western blot (C) assays revealing the expression of CUX1 and glycolytic genes in IMR32 cells stably transfected with empty vector (mock), p200 CUX1, scramble shRNA (sh‐Scb), sh‐CUX1 #1, or sh‐CUX1 #2. Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock or sh‐Scb.
-
D, E
ChIP and qPCR using Flag and CUX1 antibodies (D) and dual‐luciferase (E) assays indicating the p110 CUX1 enrichment and promoter activity of ENO1, GPI, and PGK1 in IMR32 and SH‐SY5Y cells stably transfected with mock, Flag‐tagged p110 CUX1, sh‐Scb, sh‐CUX1 #1, or sh‐CUX1 #2, and those treated with E64D (10 μM) for 24 h (n = 5). Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock, sh‐Scb, or DMSO.
-
F, G
Seahorse tracing curves (F), ECAR and OCR (G) of IMR32 cells stably transfected with mock, p110 CUX1, sh‐Scb, sh‐CUX1 #1, or sh‐CUX1 #2, and those treated with E64D (10 μM) for 24 h (n = 5). Student's t‐test, one‐way ANOVA, *P < 0.05 versus mock, sh‐Scb, or DMSO.
Data information: Data are presented as mean ± SEM. Exact
.