Figure 1.
Characterization of Cellular Heterogeneity in iPSC-CVPC Samples
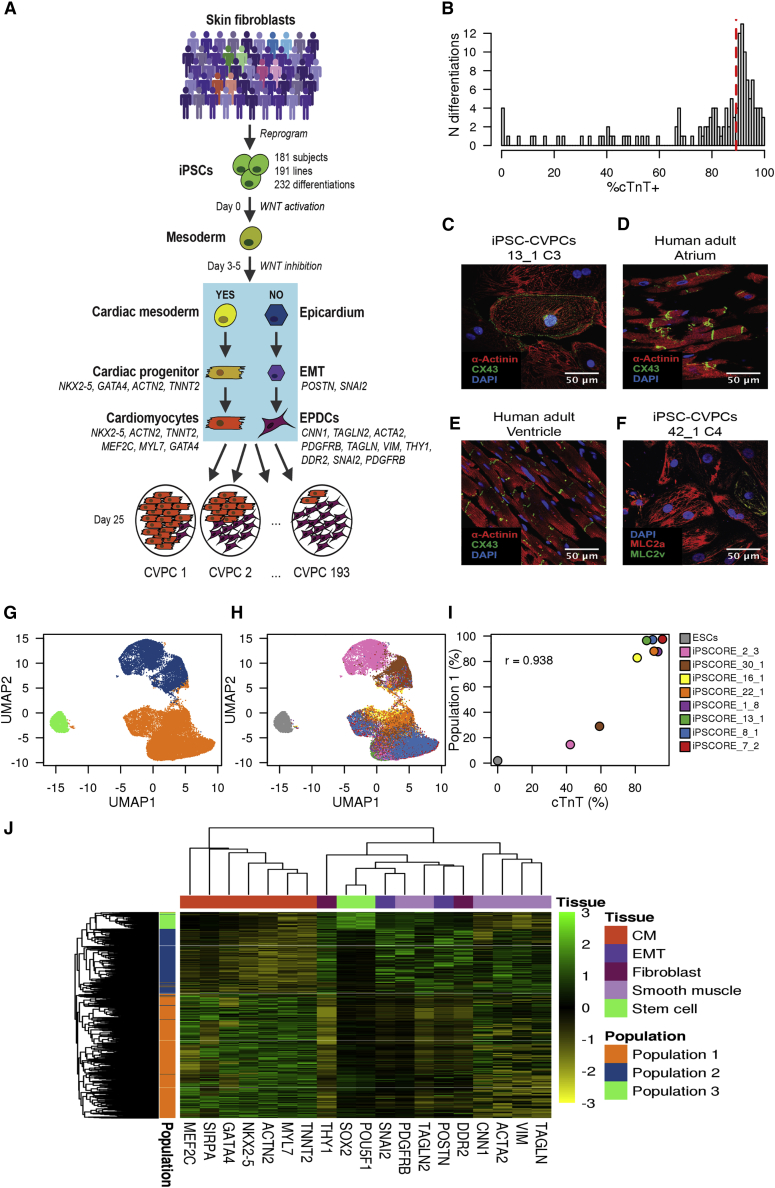
(A) Overview of the study design. Skin fibroblasts from 181 subjects were reprogrammed to iPSCs and differentiated to iPSC-CVPCs (191 lines, 232 differentiations). After WNT pathway activation at day 0 and its inactivation by IWP-2 at days 3–5, cells differentiate to CMs if WNT signaling is successfully inhibited. If WNT signaling is not sufficiently inhibited, cells differentiate to EPDCs. Of the 232 differentiations, 193 were completed (day 25), and we observed that different CVPC samples had different proportions of CMs and EPDCs.
(B) Distribution of %cTnT. Dashed red line represents the median value.
(C–E) Immunofluorescence staining of (C) iPSC-CVPCs, (D) human atrium, and (E) ventricle with markers DAPI, ACTN1, and CX43.
(F–H) Immunofluorescence staining of iPSC-CVPCs with markers DAPI, MLC2a+ and MLC2v+, and MLC2v+MLC2a+ (F). scRNA-seq UMAP plots showing (G) the presence of three populations: CMs (orange), EPDCs (blue), and ESCs (green), and (H) the distribution of the nine analyzed samples (eight iPSC-CVPC lines and one ESC line) across the three clusters.
(I) Scatterplot showing the correlation between the %cTnT and the fraction of cells in population 1 (CMs) for each of the nine samples.
(J) Heatmap showing across all 34,905 single cells the expression markers for stem cells, CMs, EMT, fibroblasts, and smooth muscle.
See also Figures S1 and S2.