Figure 4.
X Chromosome Gene Dosage Plays a Role in Cardiac Differentiation Fate
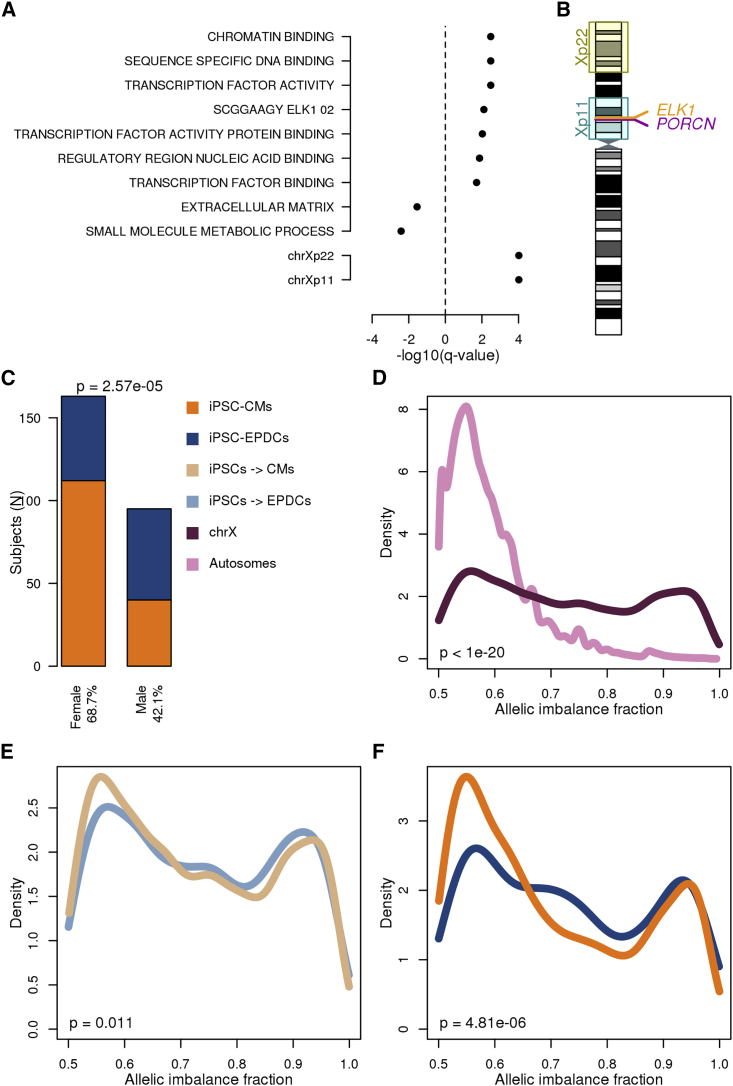
(A) GSEA results. For each gene set, −log10(q value) is shown. Positive values correspond to gene sets enriched in CM-fated iPSCs, whereas negative values correspond to EPDC-fated iPSCs. For autosomes all iPSCs were included (top), for the chromosome X only the 113 female iPSCs were analyzed (bottom). Storey q value was used to adjust for multiple testing hypothesis; q values <0.05 were considered significant.
(B) Cartoon showing the positions of differentially expressed loci on chromosome X and of ELK1 and PORCN.
(C–F) Bar plot (C) showing the associations between sex and differentiation outcome (orange: iPSC-CVPC samples with CM fraction >30%; blue: with EPDC fraction >70%). p values were calculated using Z test. Density plots showing the differences in allelic imbalance fraction between: (D) autosomal genes (pink) and chrX genes outside of the pseudoautosomal region (maroon) in female iPSCs; (E) chrX genes in female CM-fated (light orange) and EPDC-fated (light blue) iPSCs; (F) chrX genes in female day 25 iPSC-CVPC samples with CM fraction >30% (orange) and EPDC fraction >70% (blue). p values in (D) to (F) were calculated using the Mann-Whitney U test.
See also Figure S6.