Figure 2.
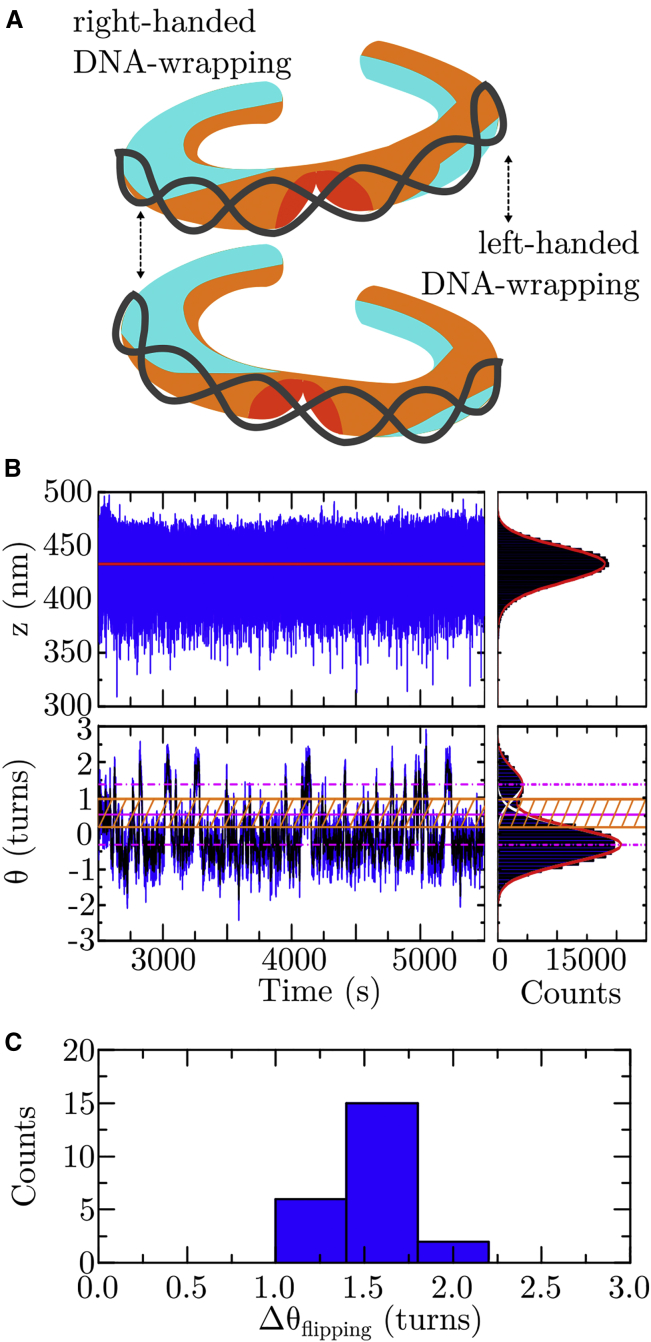
The structural dynamics of single tetrasomes on DNA with a 601 sequence. (A) Representation of the two tetrasome conformations with the DNA wrapped in either a left-handed or a right-handed superhelix. Tetrasomes were observed to spontaneously flip between these two states (35, 36, 37). This figure is adapted from (37) (https://doi.org/10.1063/1.5009100), with the permission of AIP Publishing. (B) Shown are partial time traces of the length z (in nm, top panel) and the linking number Θ (in turns, bottom panel) of a DNA molecule after the assembly of a (H3.1-H4)2 tetrasome in buffer A (Table 1) upon flushing in histone/NAP1 complexes. As indicated from the fit by the step-finder algorithm to the time trace (red line, left panel) and the fit of a mirrored γ function (red line in histogram plot, right panel) to the skewed data, the DNA length remains constant. The DNA linking number spontaneously fluctuates (i.e., “flips”) between two states identified by fitting two Gaussian functions (white lines in histogram plot, right panel) underlying the full profile (red line in histogram plot, right panel). The two states correspond to a prevalent left-handed and a less adopted right-handed conformation of DNA wrapping, with the respective mean linking numbers Θleft = −0.31 ± 0.01 turns and Θright = +1.38 ± 0.06 turns (dashed-dotted magenta lines, 95% confidence level for estimated values). Because of drift, the mean value for Θleft obtained here is offset from the average change in DNA linking number upon tetrasome dis-/assembly (Fig. 1D, bottom panel). The structural dynamics were quantified in terms of the dwell times in the two states based on a threshold zone (hatched orange area) that is bounded by 1 SD from each mean value (orange solid lines) about their average (solid magenta line) (Materials and Methods). A corresponding partial time trace of a DNAw/601 molecule upon the assembly of a tetrasome from histone tetramers only is shown in Fig. S7. (C) Shown is a histogram of the change in DNA linking number ΔΘflipping (in turns) upon flipping of single (H3.1-H4)2 tetrasomes in their handedness of DNA wrapping in all buffers (Table 1). The data yield a mean value of ΔΘflipping = 1.6 ± 0.2 turns (N = 23). The individual values are provided in Table S9. To see this figure in color, go online.