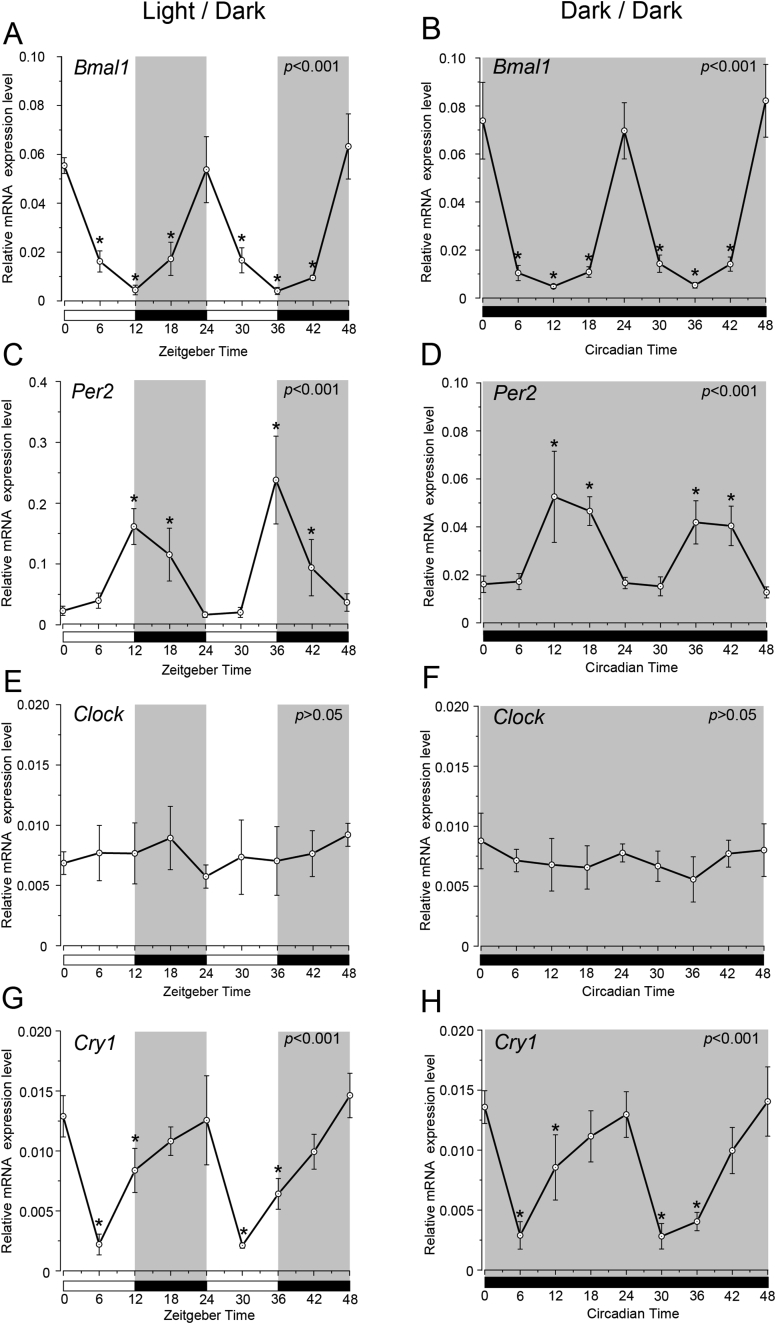
Fig. 1.
Temporal expression profiles of clock genes in the submandibular glands under LD and DD conditions. Gene expression levels of (A) Bmal1, (C) Per2, (E) Clock and (G) Cry1 at 6 h intervals in zeitgeber time (ZT: lights on at ZT0 and ZT24; lights off at ZT12 and ZT36) under the LD condition. Gene expression levels of (B) Bmal1, (D) Per2, (F) Clock and (H) Cry1 at 6 h intervals in circadian time (CT: continuous dark condition) under the DD condition. The horizontal white and black bars indicate light and dark phases (shown by gray), respectively. The mRNA levels are displayed as the mean ± SD of five replicates per time point (n = 5). P-values were calculated by one-way ANOVA and results were considered significant at p < 0.05. The Bonferroni test for post hoc comparisons was performed and p < 0.05 were considered significant differences compare to CT0 (or ZT0) are indicated as ‘*’. Rhythmicity was determined using CircWave (p < 0.05) at a 95% confidence level (α = 0.05). Bmal1, Per2 and Cry1 showed rhythmic mRNA expression patterns in both LD and DD conditions.