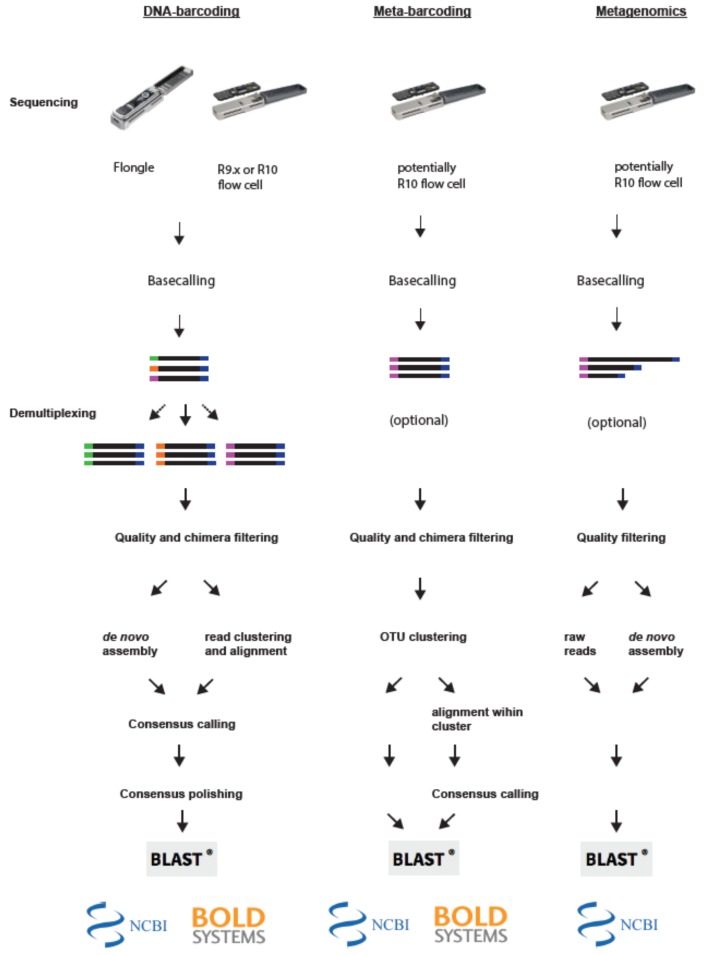
Figure 2.
Principle steps of bioinformatic pipelines for MinION-based biodiversity assessment approaches. Left panel: For DNA barcoding the reads are first base-called and demultiplexed. Next reads are filtered and depending on the applied pipeline either assembled de novo, or clustered and subsequently aligned. Lastly, the consensus sequences are polished and blasted against a database. Middle panel: For Metabarcoding applications the reads are first base-called and quality filtered. Next reads are clustered into OTUs and then blasted against a database directly or aligned to create individual consensus sequences. Right panel: For metagenomic applications reads are first base-called and filtered. Subsequently, reads are either blasted against a database directly, or assembled de novo into contigs and subsequently blasted against a database.