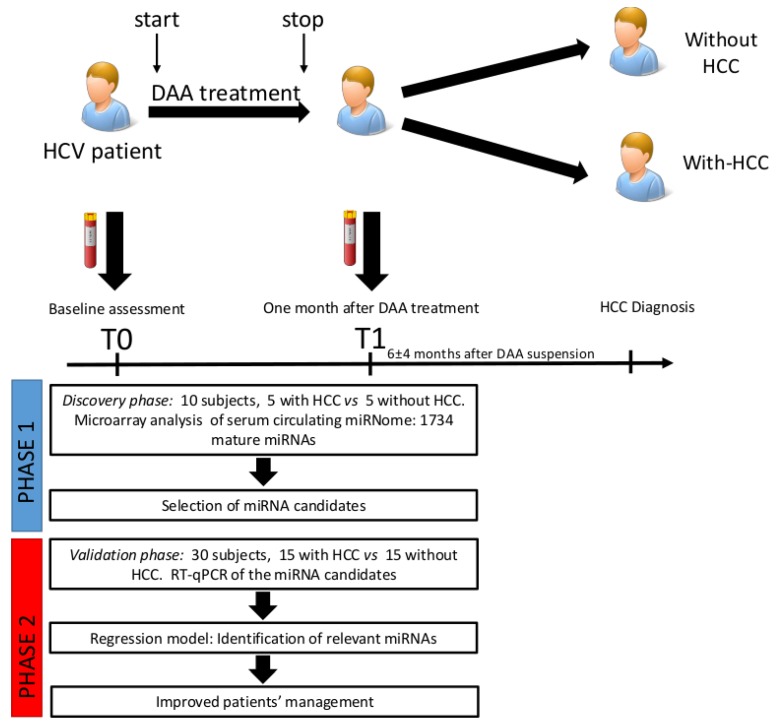
Figure 5.
General scheme of the miRNA profiling study design. In the discovery phase 10 patients were enrolled and analyzed through a Genechip miRNA 3.0 array. Patients were divided into two groups, with-HCC (subjects developing HCC after DAA treatment) and without-HCC (subjects not developing HCC after DAA treatment). Samples were analyzed at T0 and T1. Subsequently, miRNA candidates were assessed by qRT-PCR in 30 patients (validation cohort). MiRNA biomarkers were selected by estimating the time-averaged difference of a given miRNA between subjects with HCC vs. without HCC using a bootstrapped random-effect generalized least square regression model (RE-GLS).