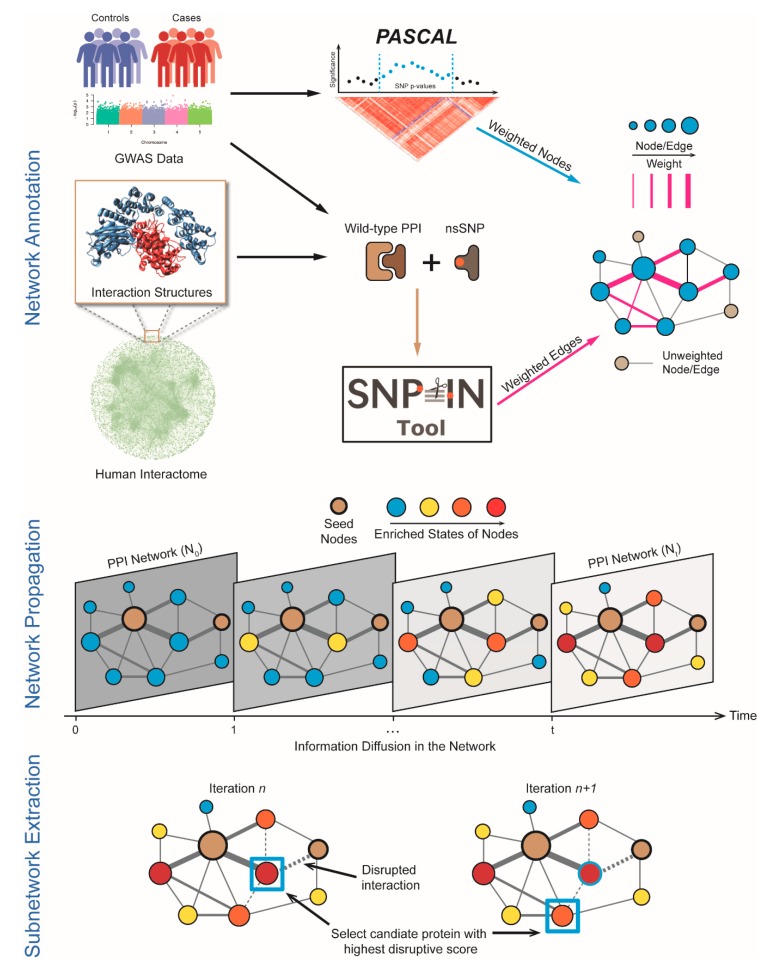
Figure 1.
Basic workflow of Discovering most IMpacted SUbnetworks in interactoMe (DIMSUM) computational framework. The module detection framework contains three major steps: network annotation; network propagation and subnetworks extraction. In Network Annotation stage, we collect GWAS data and use Pascal tool to aggregate single nucleotide polymorphisms (SNP) summary statistics. Then we apply the non-synonymous SNP INteraction effect predictor tool (SNP-IN) tool to properly assess SNP’s impact on the protein–protein interactions. Finally, we integrate this information with the human interactome to generate a fully weighted network. In Network Propagation stage, we apply a network propagation procedure to implicate genes most likely to be influenced by disruptive SNPs. Finally, in Subnetwork Extraction stage, we apply an iterative strategy to identify the most impacted module.