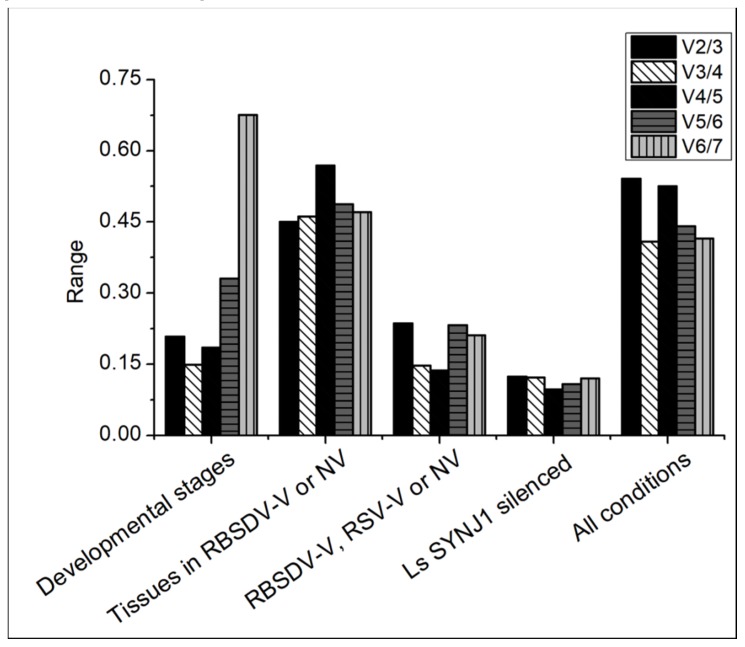
Figure 3.
Pairwise variations were calculated using the geNorm algorithm to determine the optimal numbers of reference genes needed for accurate RT-qPCR data normalization. The pairwise variation (Vn/Vn+1) was calculated using data from different developmental stages, different tissues, RBSDV-NV, RBSDV-V, or RSV-V L. striatellus, LsSYNJ1-silenced L. striatellus, and L. striatellus in all tested samples.