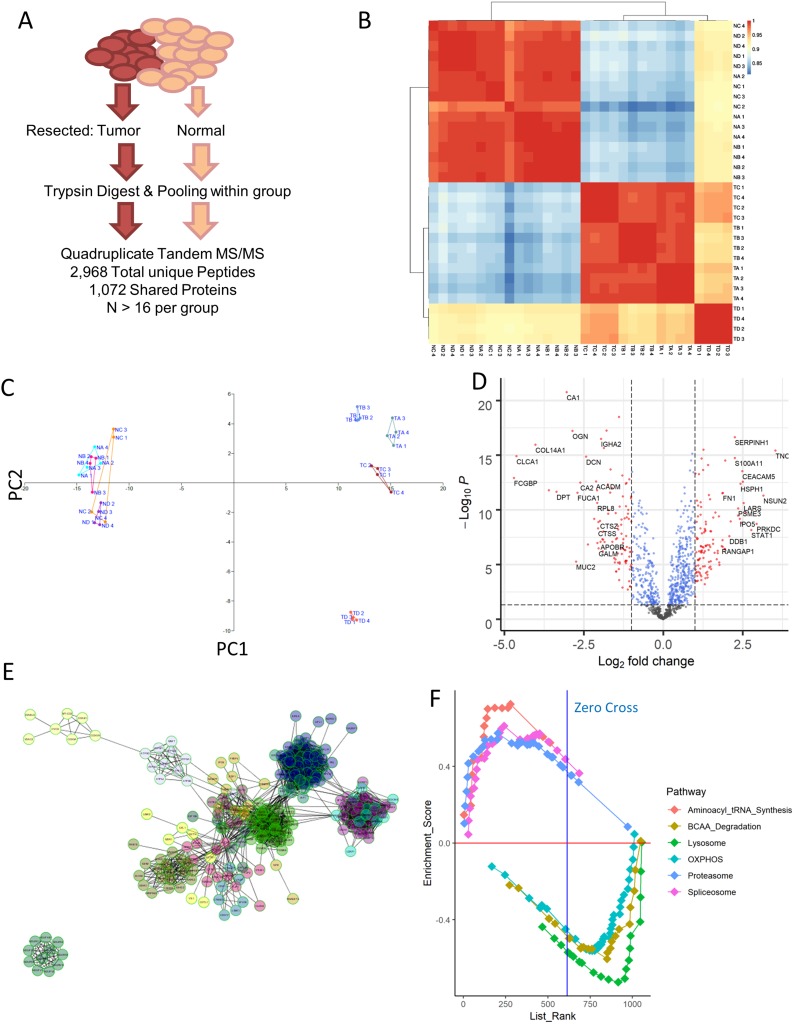
Figure 1.
Proteomic profiling of colorectal cancer. (A) Resected samples from a pool of patients were pooled together and run through mass spectrometry, yielding a collection of 2,968 peptides that match to 1071 distinct proteins shared among all the samples. (B) Correlation matrix computed using Pearson correlation between each of the samples run. NA = normal stage II, NB = normal stage III, NC = normal stage IV, TA = tumor stage II, TB = tumor stage III, TC = tumor stage IV. ND and TD samples corresponded to mucinous adenocarcinoma sample. (C) Principal component analysis of the samples confirms the stratification of tumor and normal samples seen in simple correlation analysis, while also separating out the mucinous adenocarcinoma tumor samples from the rest. (D) Volcano plot visualization of the differentially expressed genes between normal and tumor samples reveals an upregulation in some factors associated with basic cellular processes, such as PCNA and EIF3b. (E) Network analysis of proteins upregulated in tumor samples using ReactomeFI as visualized in Cytoscape identifies several classes of proteins that are highly related. (F) Gene set enrichment analysis for KEGG pathways that are differentially expressed across all normal vs tumor samples. Positive enrichment score indicates enrichment in tumor samples. The top 3 varied pathways in either direction are shown here, and are statistically significant with FDR <10% and p-value <0.05.