Fig. 5. C5 nanoparticle delivery of Cas9 RNPs enables robust CRISPR gene editing in vitro.
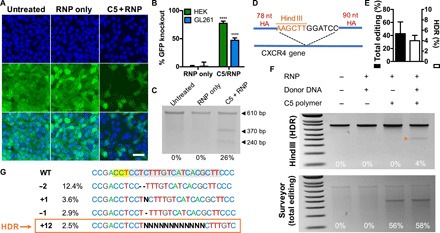
(A) Fluorescence microscopy images of HEK-GFPd2 cells treated with RNPs alone or C5 + RNPs; C5 + RNPs enabled knockout of GFP fluorescence. Scale bar, 50 μm. (B) Flow cytometry quantification of GFP knockout in HEK and GL261 cells. Data are means + SD (n = 4). Editing level of the C5/RNP group for each cell line was compared to that of the corresponding RNP-only group using Holm-Sidak corrected multiple t tests. ****P < 0.0001. (C) Surveyor mutation detector assay of GL261-GFPd2 cells treated with C5 + RNP nanoparticles. (D) Experimental design of HDR assay in the CXCR4 gene; knock-in of a 12-nt insert flanked by homology arms (HA) results in the addition of a Hind III restriction enzyme site. (E) Quantification of total editing (via Surveyor assay) and HDR (via Hind III restriction digest) in HEK cells. Data are means + SD (n = 3). (F) Hind III restriction enzyme assay (top) and Surveyor assay (bottom) of HEK cells treated with different C5/RNP/donor DNA combinations; the orange arrowhead indicates HDR. (G) Inference of CRISPR Edits analysis of Sanger sequencing data from C5 + RNP + donor DNA-treated cells provides a breakdown of different edits. Percentages indicate the percentage of the total DNA population with the indicated genotype. The targeted sequence is highlighted in gray, and the protospacer adjacent motif sequence is highlighted in yellow.
