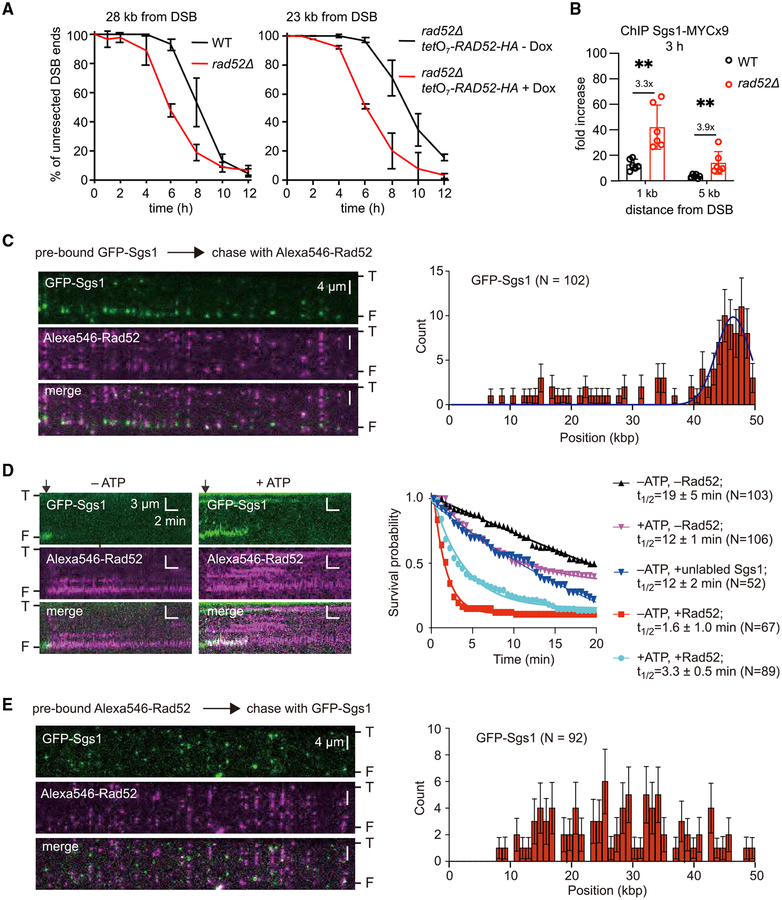
Figure 6. Rad52 Can Block DNA End Binding by Sgs1.
(A) Analysis of resection kinetics at DSB at MAT locus in budding yeast wild-type cells and cells that either lack or overexpress Rad52. Genomic DNA was digested with EcoRI; primers used to prepare DNA probes for Southern blotting are presented in Table S4. Corresponding Southern blots are shown in Figures S6A and 6B. Dox, doxycycline.
(B) ChIP-qPCR analysis of Sgs1 loading at MAT locus 3 h after DSB induction in wild-type and rad52Δ cells. Error bars denote SD (n = 6). One-tailed p values were shown (**p < 0.01).
(C) (Left) Wide-field images of GFP-Sgs1 (green) pre-bound to dsDNA ends after addition of Alexa546-Rad52 (magenta); “T” and “F” refer to the tethered and free DNA ends. (Right) Initial distribution of pre-bound GFP-Sgs1 (N = 102) that was chased by addition of Alexa546-Rad52; error bars represent 95% confidence intervals (CIs), and the blue line represents a Gaussian fit to the data.
(D) (Left) Kymograph showing the dissociation of end-bound GFP-Sgs1 (green) after addition of Alexa546-Rad52 (magenta). Arrowheads denote the time point of the Alexa546-Rad52 injection. (Right) Survival probability for end-bound GFP-Sgs1 (±ATP) chased with Rad52 or unlabeled Sgs1, as indicted. Note, that the minus Rad52 data curves are reproduced from Xue et al. (2019) for comparison.
(E) (Left) Wide-field images showing dsDNA pre-bound by Alexa546-Rad52 (magenta) and then chased with GFP-Sgs1 (green). (Right) The corresponding GFPSgs1 binding distribution. Error bars represent 95% CIs.