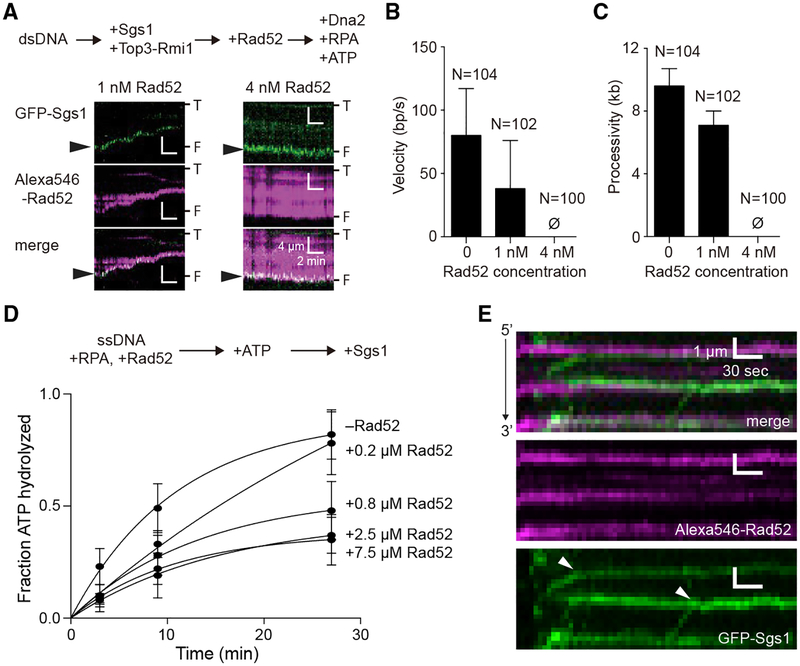
Figure 7. Rad52 Inhibits the Motor Activities of Sgs1.
(A) Kymographs depicting GFP-Sgs1 (green) during dsDNA end resection assays on DNA pre-bound by Alexa546-Rad52 (magenta). Arrowheads highlight the position of GFP-Sgs1 at the DNA ends at the beginning of the measurement. “T” and “F” refer to the tethered and free ends of the DNA.
(B) Velocity of the dsDNA resection machinery (GFP-Sgs1, Top3-Rmi1, Dna2, and RPA) in assays containing either no Rad52, 1 nM or 4 nM Alexa546-Rad52, as indicated. Error bars represent SD obtained from Gaussian fits to the data.
(C) Processivity of the dsDNA resection machinery (GFP-Sgs1, Top3-Rmi1, Dna2, and RPA) in assays containing no Rad52 or 1 nM or 4 nM Alexa546-Rad52, as indicated. Error bars represent SD obtained from Gaussian fits to the data.
(D) Sgs1 ATP hydrolysis activity in the presence of Rad52. Error bars represent the SD of three independent experiments.
(E) Kymographs depicting the translocation activity of GFP-Sgs1 (green) on RPA-ssDNA (unlabeled) in the presence of Alexa546-Rad52 (magenta). White arrowheads in the GFP-Sgs1 image illustrate the positions at which translocating molecules of GFP-Sgs1 terminate translocation upon encountering Alexa546-Rad52. Note that the minus Rad52 data columns in (B) and (C) are reproduced from Xue et al. (2019) for comparison.