Figure 3: Loss of Prdm16 in stem cells leads to TA cell apoptosis and diminished epithelial differentiation.
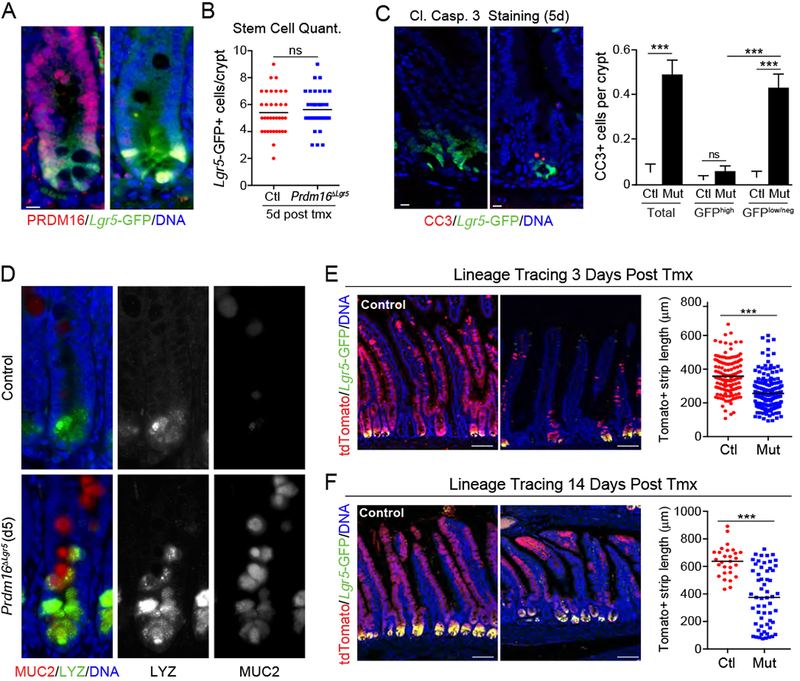
A-D) Lgr5GFP/Cre-ERT2;Prdm16loxP/loxP mice were treated with tmx (mutant; Prdm16ΔLgr5) or vehicle (corn oil; Control). A) Immunostaining of PRDM16 (red) and Lgr5-GFP (green) in duodenal crypts at 7d post tmx. DNA (DAPI, blue). B) Quantification of Lgr5GFP+ stem cells per crypt at 5d. n=35 crypts from 3 mice. C) (left) Duodenal crypts stained for cleaved Caspase 3 (CC3; red) at 5d. (right) Quantification of CC3+ stem (GFPhi) or TA (GFPlow/neg) cells using an intensity threshold (Thermal LUT in Image J). Controls were shared with experiment in Figure 2B. n=91-188 crypts from 3 mice per group. D) Immunostaining of Mucin2 (MUC2; red) and Lysozyme (LYZ; green) in duodenal crypts at 5d. Arrowheads indicate MUC2/LYZ double-positive cells. n=3 mice.
E-F) Lineage tracing of Lgr5+ stem cell progeny in Control (Prdm16loxP/+) and Prdm16 mutant (Prdm16loxP/oxP; Prdm16ΔLgr5) reporter mice (Lgr5GFP/Cre-ERT2;R26Rlox-stop-lox-tdTomato). Marked epithelial strip length was measured for continuous streaks of tdTomato+ (Red) epithelial cells ascending the villi at: E) 3d [n=132-190 villi from 3 mice] or F) 14d [n=28-58 villi from 3 mice] post tmx. Lgr5-GFP (green), DNA (DAPI, blue).
All panels show mean ± SEM, *p<0.05, **p<0.01, ***p<0.001. Scale bars: 10 μm (A, C, ; (D) 100 μm (E, F).
