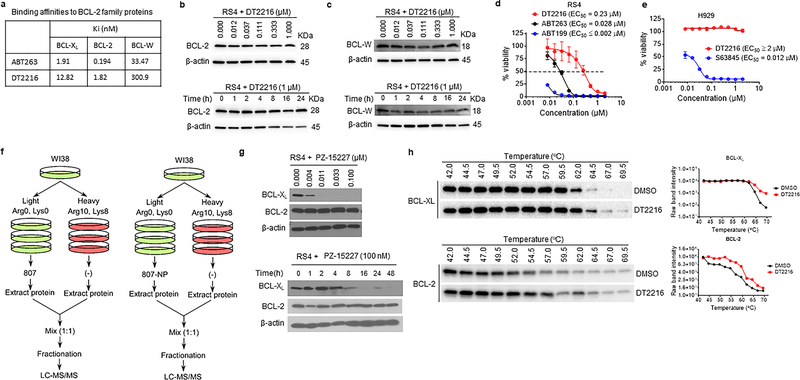
Extended Data Fig. 4. DT2216 binds to both BCL-XL and BCL-2 but cannot degrade BCL-2.
a, The binding affinities of DT2216 and ABT263 towards BCL-XL, BCL-2 and BCL-W were measured by AlphaScreen and are represented in terms of inhibition constant (Ki). The data are average of two independent experiments each performed in duplicates. b, c, Immunoblot analyses of BCL-2 and BCL-W are shown after the RS4 cells were treated with indicated concentrations of DT2216 for 16 h (upper panels), or with 1 μM of DT2216 for indicated durations (lower panels). d, Cell viability of BCL-2-dependent RS4 cells after treatment with increasing concentrations of DT2216, ABT199 or ABT263 for 72 h. Data are presented as mean ± SD from six replicate cell cultures in one experiment. Similar results were obtained in two additional independent experiments for DT2216. The EC50 of DT2216 is the average of three independent experiments. e, Cell viability of MCL-1-dependent H929 cells after treatment with increasing concentrations of DT2216 or S63845 for 72 h. Data are presented as mean ± SD from six replicate cell cultures in one experiment. f, Schematic representation of proteomic assay shown in fig. 3d. g, Representative immunoblot analyses are shown after the cells were treated with indicated concentrations of PZ-15227 for 16 h (top panel) or with 0.1 μM of PZ-15227 for indicated time points (bottom panel). Similar results were obtained in one more independent experiment. h, CETSA assay for BCL-XL and BCL-2. MOLT-4 (for BCL-XL) or RS4 (for BCL-2) cells were treated with DMSO, or 1 μM of DT2216 for 6 h. Raw band intensities are mean of two independent experiments.