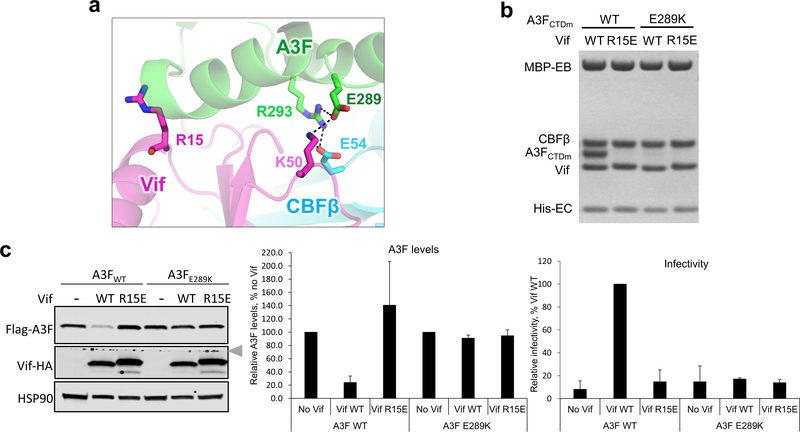
Extended Data Fig. 7. HIV1-Vif R15 does not interact with A3F E289.
a, Vif R15 is located far away from A3F E289, which interacts with Vif K50 in the Vif–CBFβ–A3FCTDm structure. b, Mutational analysis of the interactions by in vitro binding assay using MBP-tagged Vif–CBFβ–EloB–EloC variants to pulldown A3FCTDm variants. The loading controls are shown in Supplementary Fig. 1b & c. The charge-swapped Vif R15E/A3F E289K double mutation did not rescue the Vif-A3F interaction in vitro. c, The effect of Vif R15E or A3F E289K mutation on A3F sensitivity to Vif-mediated degradation indicated by western blot (left), quantified A3F levels relative to No Vif (mean ± s.d.; n=3 biologically independent experiments; middle), and relative viral infectivity (mean ± s.d.; n=3 biologically independent experiments; right). In contrast to the prior report 37, the charge-swapped Vif R15E/A3F E289K double mutation did not restore the Vif-mediated A3F degradation or viral infectivity in cells. The blot was cut as indicated (gray arrow), where one half was used to detect Flag-A3F and HSP90, and the other half was used to detect Vif-HA.