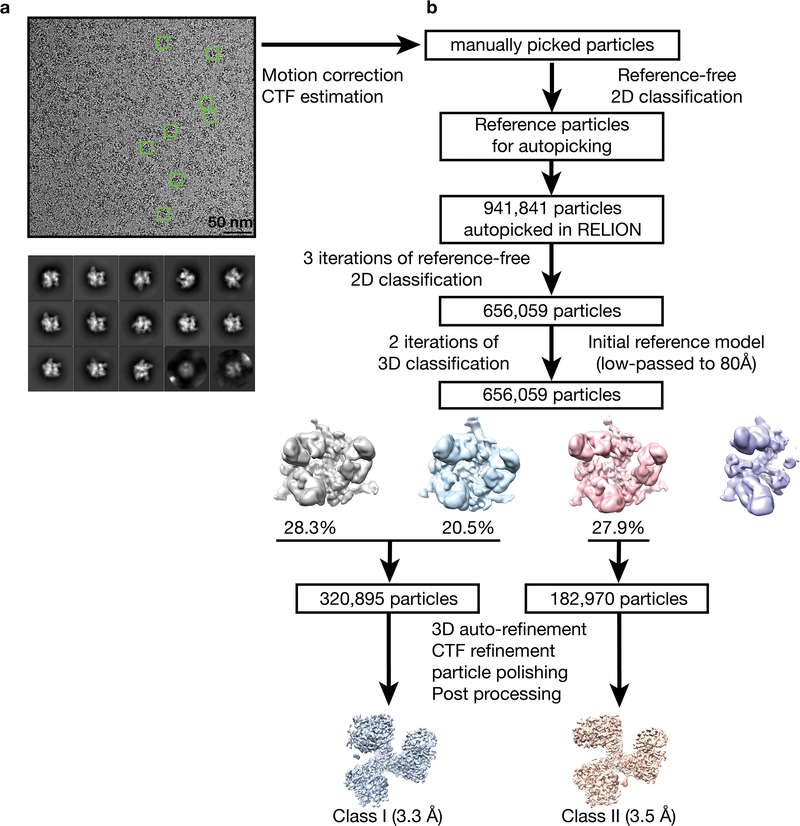
Extended Data Fig. 2. Data processing for BG505 SOSIP-sCD4-E51 complex structures.
a, Example of a motion-corrected and dose-weighted micrograph of E51-sCD4-BG505 complexes (representative individual particles in boxes). Scale bar = 50 nm. Defocus was ~2.6 μm underfocus. b, Data processing scheme. Collected micrographs were motion-corrected using MotionCor2 (ref.1) without binning. Contrast transfer functions (CTFs) were estimated using Gctf 1.06 (ref.2). ~1,000 particles were manually picked and subjected to 2D classification in RELION3,4. Particles from good classes were used as references for automatic particle picking using RELION AutoPicking. 941,841 autopicked particles were subjected to three rounds of reference-free 2D classification. In each round, particles from good 2D classes were selected, which gave 656,059 particles. Subsequently, using the Env trimer portion of an 80 Å low-pass filtered partially-open trimeric BG505 SOSIP structure (PDB 6CM3) as the reference model, first round of 3D classification was performed assuming C1 symmetry. Particles from good classes were combined and used for a second round of 3D classification with C1 symmetry. The resulting maps could be grouped into two classes: class I (320,895 particles) and class II (182,970 particles). Maps from the two classes were refined separately assuming C1 symmetry with the sCD4 D2 domain and E51 Fab CHCL domains masked out. After CTF refinement, movie refinement, and particle polishing, the class I and class II post-processed maps were refined to 3.3 Å and 3.5 Å resolution (FSC 0.143), respectively (Fig. S3).