Figure 2.
Differential Promoter-Enhancer Interactions after RUNX1-ETO Depletion Are Driven by Differential TF Binding
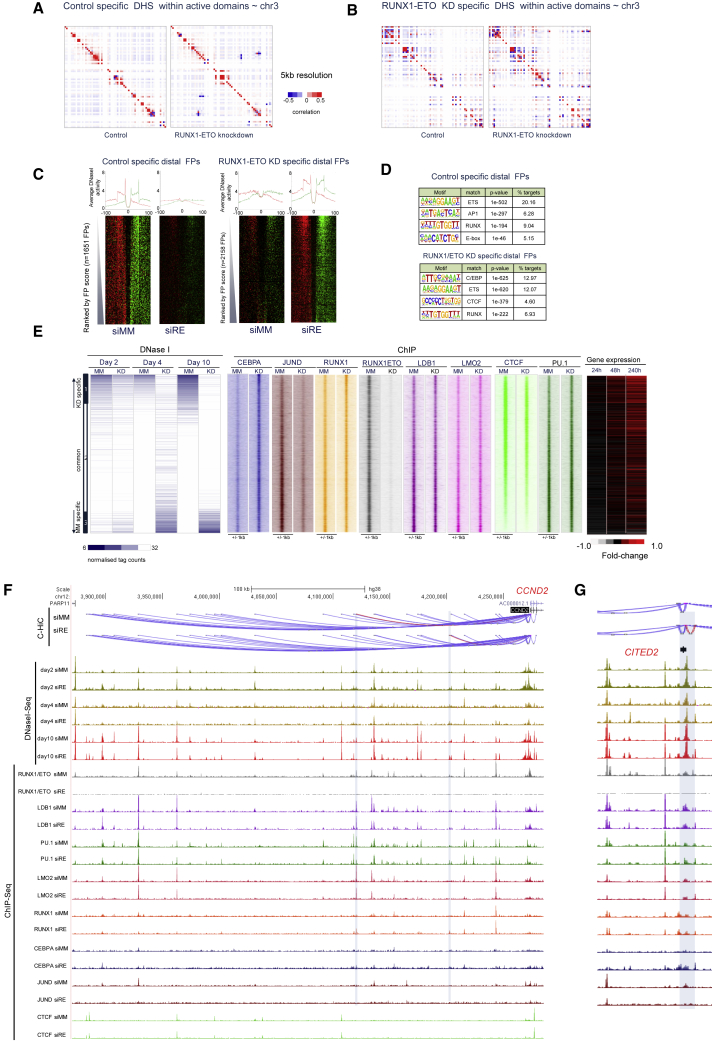
(A) Heatmap representing the correlation of normalized interaction ratios across chr3 at 5-kb resolution, showing the correlation of CHiC peaks in regions specific to DHS peaks that are depleted after RUNX1-ETO knockdown. Each pixel represents a 5-kb section of the genome. The left panel shows the interaction heatmap for siMM and the right panel for siRE cells. Positive correlations are shown as red; negative correlation as blue squares. To determine statistically significant interactions, reads from replicates 1 and 2 were merged.
(B) Heatmap representing the correlation of normalized interaction ratios across chr3 at 5-kb resolution and showing the correlation of CHi-C peaks in DHS peaks that are newly formed after RUNX1-ETO (R/E) gene knockdown. For all other features, see (A).
(C) DNaseI cleavage patterns within specific distal footprints predicted by Wellington (Piper et al., 2013). Upper strand cut sites are shown in red and lower strand cut sites in green within a 200-bp window centered on each footprint (gap) for siMM- and siRE-specific footprints.
(D) Analysis of overrepresented binding motifs within each footprint class as defined in (C).
(E) Left panel: time course of DHS development after 2, 4, and 10 days of RUNX1-ETO depletion (see scheme in Figure S1E). Normalized tag counts are ranked alongside day-10 knockdown (KD) and control-specific (bottom) counts; common and siRE-specific DHS are indicated on the left. Alongside the same genomic coordinates, C/EBPα, JUND, LDB1, CTCF, RUNX1-ETO, LMO2, PU.1, and RUNX1 ChIP-seq reads from Kasumi-1 cells with or without RUNX1-ETO depletion are plotted as indicated (middle panel). The right panel shows the expression levels of the genes linked to the associated DNaseI-seq sites (right panel).
(F) UCSC browser screenshot depicting interactions between the CCND2 promoter and surrounding DHS (shown as arcs) together with the indicated ChIP-seq data before and after RUNX1-ETO knockdown. Changing interactions are shown in red, and their associated DHS/ChIP peaks are highlighted using a vertical shaded bar.
(G) The same analysis as in (F) for the CITED2 locus.