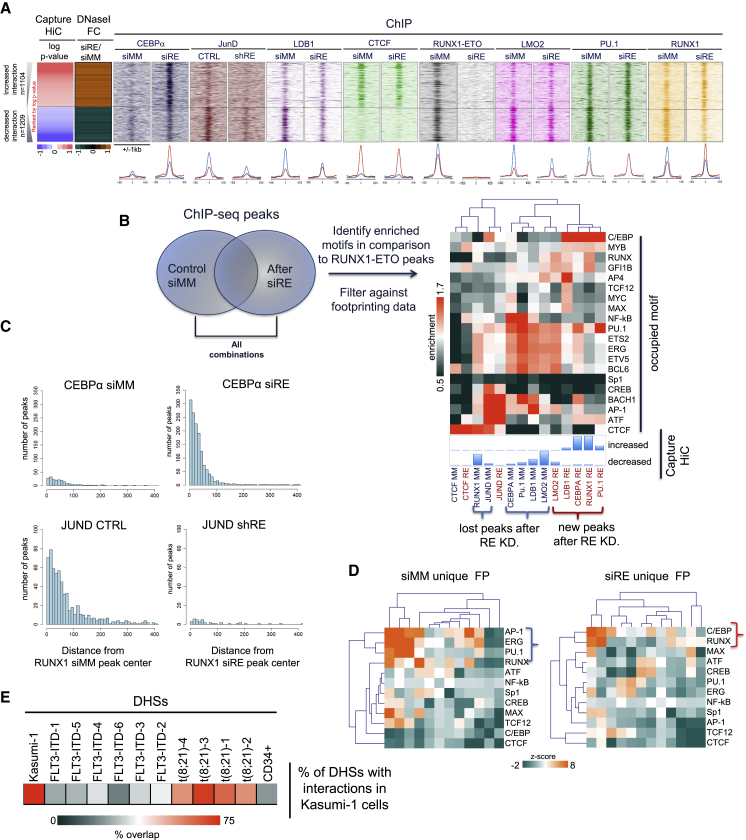
Figure 3.
The Cooperation of Constitutive and Inducible TFs Is Associated with Differential Interactions
(A) Log p values of the differential interactions were plotted ranked from high to low for control and RUNX1-ETO-depleted cells. Red represents an increase in interaction strength and blue represents a decrease. Alongside, the DNaseI-seq fold difference between control and RUNX1-ETO knockdown cells as well as ChIP-seq density profiles for C/EBPα, JUND, LDB1, CTCF, RUNX1-ETO, LMO2, and PU.1 are plotted from Kasumi-1 cells, transfected with either siMM or siRE as indicated. The panels below show the average profiles of the binding of the indicated TFs plotted around the peak summit for control and RUNX1-ETO-depleted cells. Red, ChIP signal specific for peaks with increased interactions; blue, ChIP signal specific for peaks with decreased interactions.
(B) Determination of enriched motifs for other TFs in ChIP-seq peaks specific for control and RUNX1-ETO-depleted cells. Motif enrichment was first identified using HOMER and then filtered against digital footprinting data from day 10 of knockout to ensure that these binding motifs were functional. Enrichment scores were subjected to unsupervised clustering for each of the indicated motifs (on the right). The heatmap depicts the degree of motif enrichment with highly enriched motifs shown in red. Peaks were overlaid with the DHS that show new interactions (red brackets at the bottom) or whose interactions are lost (blue brackets). Enrichment scores were calculated by the level of motif enrichment in the unique peaks, as compared to motif enrichment in RUNX1-ETO peaks. Bottom panels: percentage of peaks showing differential interaction with TFs binding to these sites as determined by ChIP-seq (control cells, blue; RUNX1-ETO-depleted cells, red).
(C) Bar plots illustrating the distribution of distances between the binding sites of the indicated TFs as determined by ChIP-seq. We measured the changing distance between RUNX1 peaks in siMM and siRE cells and C/EBPα peaks in siMM (top left) and siRE cells (top right), as well as the distance between RUNX1 peaks and JUND control peaks (bottom left) and JUND after R/E KD (bottom right).
(D) Bootstrapping analysis of the significance of co-localizing of footprinted motifs within day-10 DHSs for sites that are either lost (left panel) or gained (right panel) after RUNX1-ETO depletion as compared to the rest of the genome. The heatmap shows the significance of motifs co-localizing within 50 bp as compared to sampling by chance.
(E) Heatmap highlighting the percentage of day-4 Kasumi-1 DHSs with interactions found in different patient groups indicating the similarity between cell-line and primary t(8;21) data. The t(8;21) and FLT3-ITD DHS/CHi-C patient data were downloaded from GEO: GSE108316 (Assi et al., 2019).