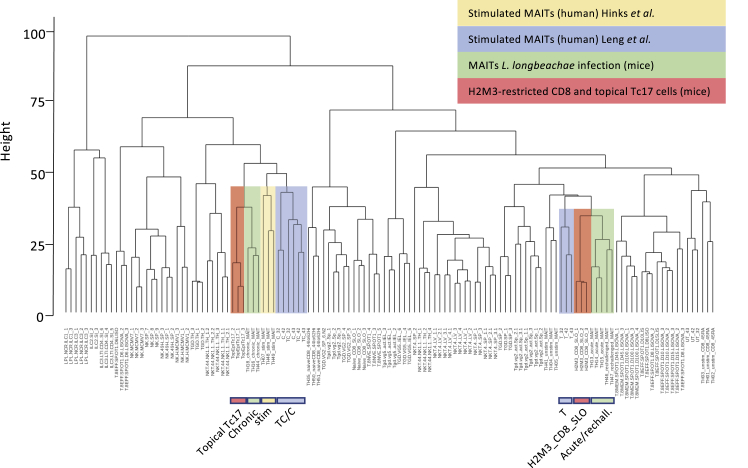
Figure 6.
Integrated Transcriptional Analyses Reveal the Relationship between In Vitro-Activated Human MAIT Cells and In Vivo-Activated Murine MAIT and Tc17 Cells
Hierarchical clustering analysis of the transcriptomic profiles of the indicated cell populations is shown. Similarity between the expression profiles is measured using a Euclidean distance (height). Datasets were derived from ImmGen (Heng et al., 2008), Linehan et al. (2018), and Hinks et al. (2019) and were integrated as described in the STAR Methods section. The relevant datasets are colored. UT, TCR, C, and TC refer to the conditions used in this paper on in vitro-activated human CD8+ MAIT cells (blue). Topical Tc17 and H2M3_CD8_SLO refer to the H2M3-restricted populations identified in Linehan et al. (2018) in the skin and secondary lymphoid organs of mice, respectively (red). The cells described in Hinks et al. (2019) are marked in yellow (stimulated human MAIT cells) or green (chronic or acute, derived during a late or an early time point after L. longbeachae infection in mice, respectively). See also Figure S6.