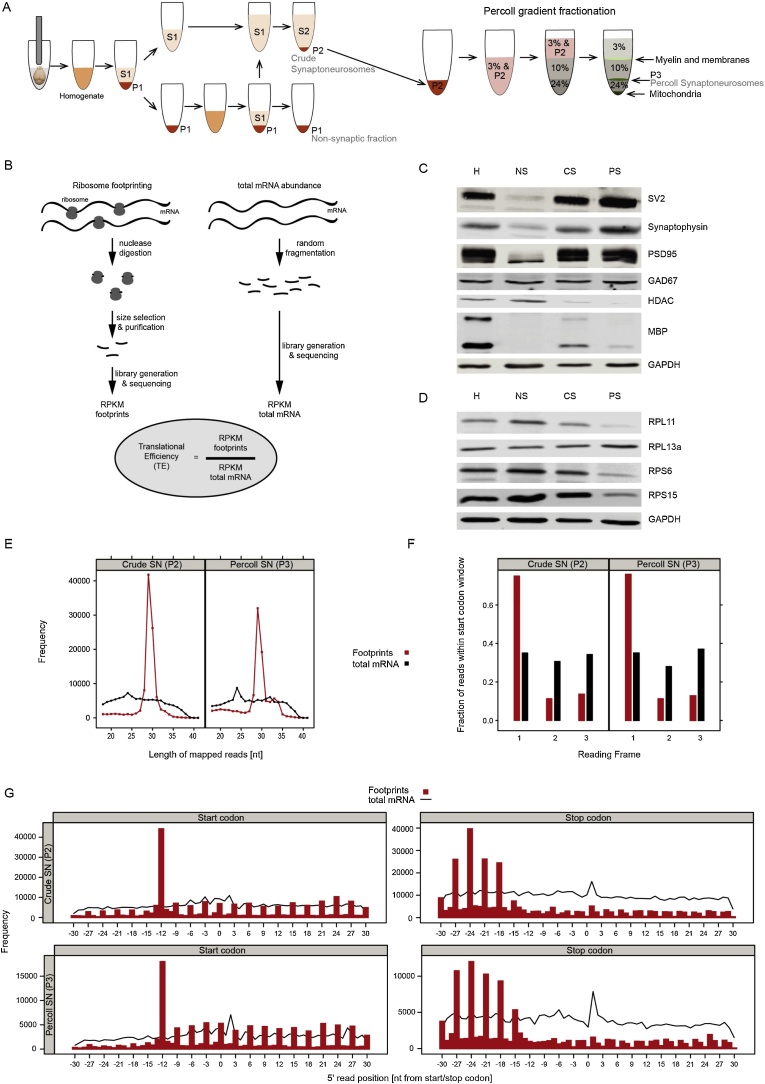
Fig. 1.
Schematic diagram of the preparation of SN (A) and the ribosome profiling workflow (B). A detailed description can be found in the Methods section. (C) Immunoblots of the indicated proteins in the different fractions obtained during the preparation of SN. Note the depletion of nuclear proteins (HDAC) and the enrichment in both excitatory and inhibitory synaptic proteins (synaptic vesicle glycoprotein 2A - SV2, synaptophysin, glutamic acid decarboxylase 67 – GAD67, and postsynaptic density protein 95 - PSD95) in crude and Percoll SN. PSD95 blots show two bands very close in size, representing α and ß isoforms of the protein, respectively (Chetkovich et al., 2002). In addition, Percoll SN show a major decrease in myelin components (MBP) of both major isoforms. GAPDH was used as a loading control. H: tissue homogenate, NS: non-synaptic fraction, CS: crude synaptoneurosomes, PS: Percoll synaptoneurosomes. (D) Immunoblots confirming the presence of ribosomal proteins (large ribosomal proteins 11 and 13a, small ribosomal proteins 6 and 15) in the SN fractions. GAPDH was used as a loading control. (E) Size distribution of the aligned sequencing reads from the indicated samples. Total mRNA reads show a random size distribution, whereas ribosomal footprints show a distinct peak between 28 and 30 nt. (F) Reading frame usage in the total mRNA and footprint samples, showing the preferential alignment within the first reading frame in the footprint samples, compared to the total mRNA samples which have been randomly fragmented. (G) The total number of read fragments aligning around the start and stop codons of the coding sequence of all genes. Footprints show a 3 nt periodicity, compared to total mRNA reads.