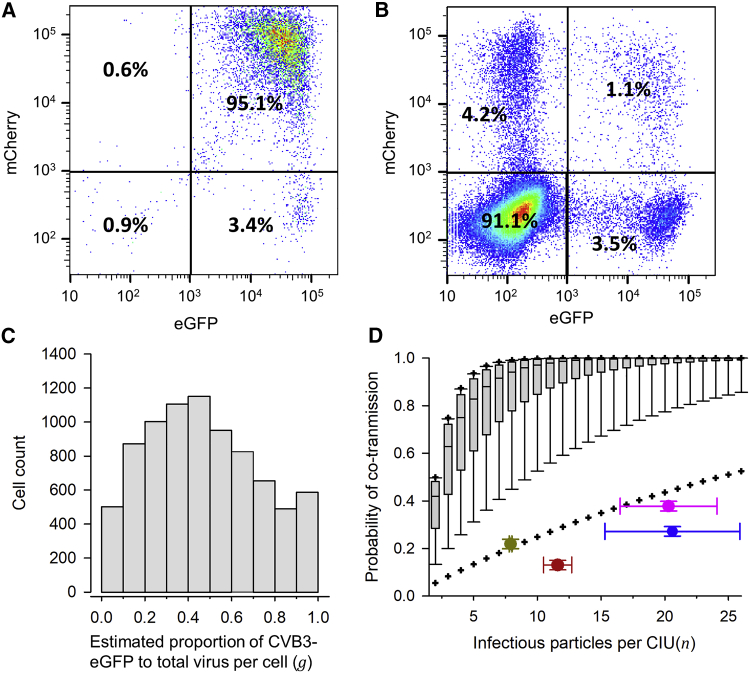
Figure 2.
Infrequent Co-transmission of EGFP- and mCherry-Encoding CVB3 Viruses through CIUs
(A) Flow cytometry analysis of producer cells inoculated at high density (D ≈ 10 PFUs/cell) with a 1:1 mix of the two variants (culture media harvested at 12 hpi).
(B) Flow cytometry analysis of cells inoculated at lower density (D ≈ 0.1 PFU/cell) with the P3 fraction. Results for one replicate are shown. Cell counts for three replicates are shown in Table S1.
(C) Distribution of the estimated proportion of each variant in individual producer cells (g), rescaling the intensities shown in (A) such that the average rescaled intensities matched the observed proportion of cells infected with each variant (46.4% ± 0.4% CVB3-EGFP versus 53.6% ± 0.4% CVB3-mCherry; Table S1). This was achieved by multiplying all EGFP intensities by 1.82. The resulting coefficients of variation of mCherry and rescaled EGFP intensities were similar (0.72 and 0.68, respectively).
(D) Boxplot of the probability that CIUs containing n infectious particles co-transmitted both variants, assuming random mixing. This probability was calculated as p(n) = 1 − gn − (1 − g)n. The lower and upper limits of the box indicate 25th and 75th percentiles, and the middle line shows the median. Whiskers show the 10th and 90th percentiles, and crosses the 5th and 95th percentiles. Superimposed are shown the n value and observed proportion of coinfected cells in four different cases: P3 fraction harvested at 12 hpi (red), P3 fraction harvested at 12 hpi accounting for possible contamination with free virions (pink), P3 fraction harvested at 8 hpi (brown), and P2∗ fraction harvested at 12 hpi (blue; see text for details). The mean and SEM from three independent assays are shown.