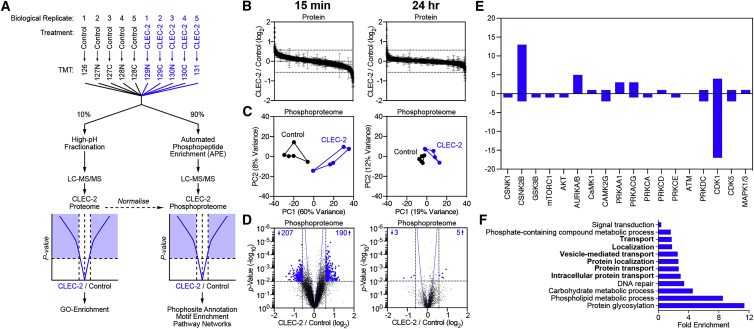
Figure 3.
Phosphoproteomics of CLEC-2-Fc-Treated FRCs
(A) Experimental setup, comparison of 5 control (untreated) and 5 CLEC-2-Fc-treated FRC cell lysates (biological replicates).
(B) Waterfall plots showing proteome regulation by CLEC-2-Fc.
(C) Control and CLEC-2-Fc-treated phosphoproteomes cluster in a PCA space.
(D) Volcano plots showing statistical regulation of the CLEC-2-Fc-treated FRC’s phosphoproteome (n = 5, two-tailed t test, Gaussian regression). The number of regulated phosphosites is indicated.
(E) Empirical parent kinase analysis. The bars represent the number of targets for kinases. Positive and negative values indicate higher or lower phosphorylation in CLEC-2-Fc-treated cells.
(F) Gene Ontology analysis for biological processes. Each bar represents a biological process significantly enriched by binomial analysis.