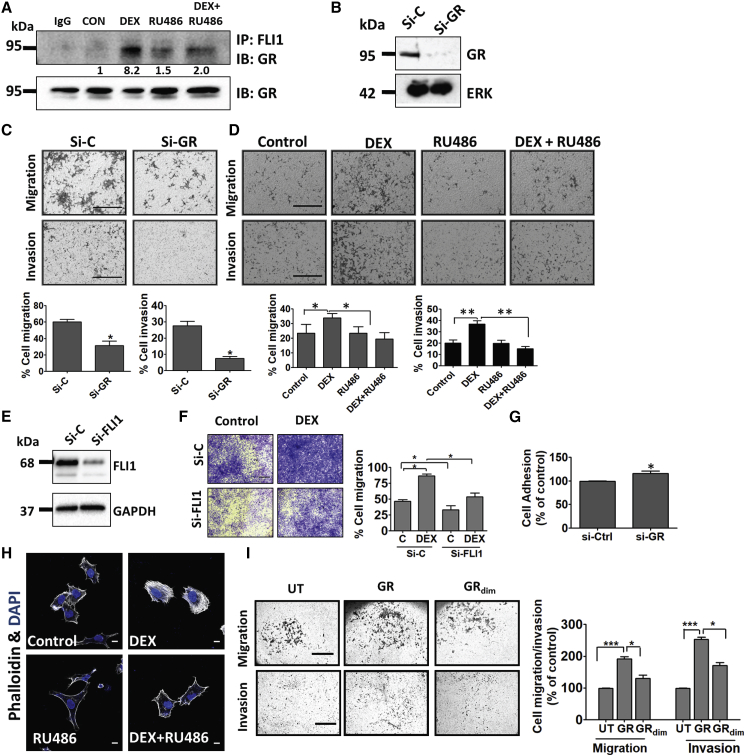
Figure 4.
Ligand-Induced Activation of GR Associates with Increased Migration and Invasion of Ewing Sarcoma Cells
(A) Serum-starved CHLA9 cells were treated (60 min) with vehicle, DEX (1 μM), and RU486 (1 μM). Extracts were processed for co-immunoprecipitation (IP) and immunoblotting (IB). Images are representative of 3 replicates. Signals were quantified and normalized. IgG, control rabbit antibody.
(B) CHLA9 cells were transfected with either GR-specific or control-scrambled siRNAs (si-C). Knockdown efficiency was tested after 48 h using immunoblotting with antibodies to GR.
(C and D) CHLA9 cells were seeded in Transwell migration chambers or Matrigel-coated invasion chambers. (C) Control siRNAs or siRNAs specific to GR were added 24 h prior to seeding, and both migration and invasion were measured 20 h later or, alternatively, (D) DEX (1 μM) and RU486 (1 μM) were added, and migration and invasion were assayed. ∗p ≤ 0.05, ∗∗p ≤ 0.01. Bars, 500 μm.
(E) CHLA9 cells were transfected with either FLI1-specific or control siRNAs (si-C), and knockdown efficiency was tested 48 h later.
(F) CHLA9 cells were treated with siRNAs as in (E). Twenty-four h later, cells were seeded in Transwell migration chambers and incubated for 20 h with DEX. Thereafter, cell migration was quantified. ∗p ≤ 0.05. Bars, 500 μm.
(G) Plates were pre-coated with fibronectin, and then CHLA9 cells (pre-transfected with si-GR or si-C) were seeded and allowed to attach for 90 min. Adherent cells were stained and optical density (550 nm) was quantified in triplicates. ∗p < 0.05.
(H) A673 cells were incubated for 24 h with either DEX or RU486 and thereafter fixed and stained with DAPI and phalloidin. Bar, 10 μm.
(I) CHLA9 cells were transfected with vectors encoding GR, GRdim− (A458T), or they were un-transfected (UT). Twenty-four h later, we assayed migration and invasion. Signal quantification (means ± SD) and representative fields are presented. ∗p ≤ 0.05; ∗∗∗p ≤ 0.001. Bars, 500 μm (migration) or 100 μm (invasion).