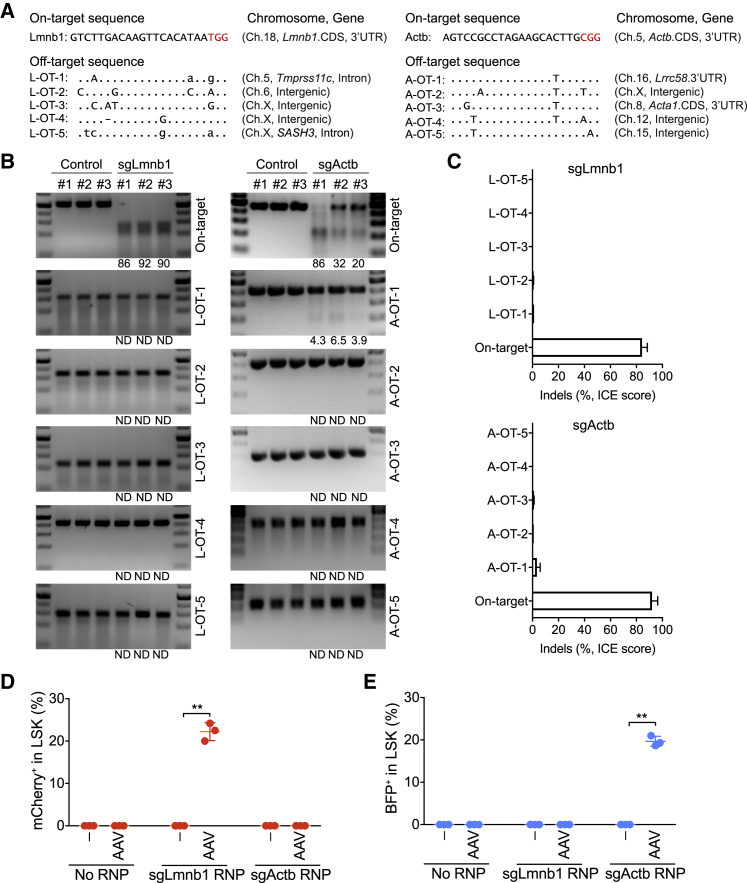
Figure 2.
Off-Target Analysis in Mouse HSPCs
(A) On-target and 5 highest-risk off-target sites of sgLmnb1 and sgActb as predicted by CrispRGold.
(B) T7EI assays show editing activity of sgLmnb1 (left) and sgActb (right) in the on- and/or off-target sites from control or treated HSPCs (ND, not determined).
(C) ICE analysis showing the indel frequency in the on- and/or off-targeted sites. Data are shown as means ± SD from three independent experiments.
(D and E) Activated-Sca1+ HSPCs from C57BL/6 mice were electroporated with RNPs containing the indicated sgRNAs and infected with the indicated donor viruses. The frequency of mCherry+ (D) and BFP+ (E) LSK cells was determined by flow cytometry based on three independent experiments. Data are shown as means ± SD (∗∗p < 0.01, Mann-Whitney test).