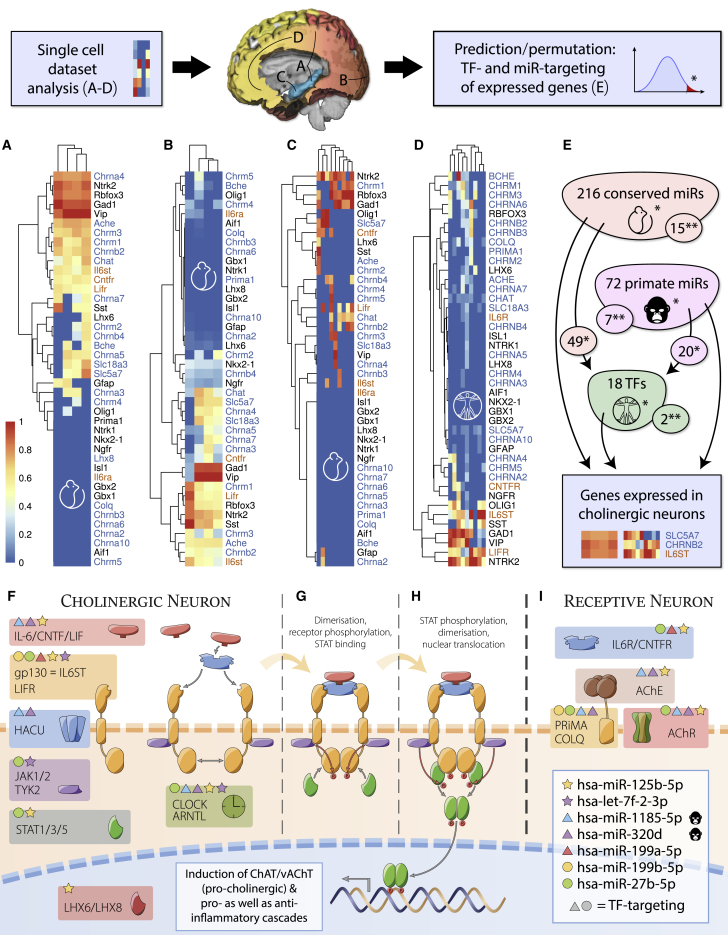
Figure 3.
Single-Cell Sequencing of ChAT/vAChT-Positive Cortical Cells and Analysis of Their Expressed Transcripts
Expression values were normalized (0–1) for each dataset. The order of genes in each heatmap reflects transcript clustering rather than level differences. Columns represent individual samples from original data (see column names in Figure S1); cholinergic genes are denoted in blue and neurokine receptors in orange.
(A) Clustered single-cell sequences of the transgenic mouse somatosensory cortex and hippocampus (Zeisel et al., 2015).
(B) Clustered single-cell sequences from the transgenic mouse visual cortex (Tasic et al., 2016).
(C) Single-nucleus sequencing of adult mouse hippocampus (Habib et al., 2016).
(D) Single-cell sequencing of the human developing neocortex (Darmanis et al., 2015).
(E) Permutation analysis of miRNA and TF targeting data of genes expressed in cholinergic cells (via miRNet) identified putative cholinergic/neurokine co-regulators (∗p < 0.05, ∗∗p < 0.001). Implicated TFs are regulated by a subset of miRNAs targeting cholinergic genes, indicating nested regulation (details in Data S2).
(F–I) Gp130-family neurokine, cholinergic, and circadian signaling pathways are controlled by primate-specific and evolutionarily conserved miRNAs. Components expressed in cholinergic neurons (F), processes following receptor binding (G) and STAT phosphorylation (H), and components of cholinoceptive neurons (I) are shown. miRNA targeting of individual genes (indicated by colored symbols) yields complex transcriptional interactions. Several miRNAs directly targeting the cholinergic pathway also target TFs controlling this pathway (circles and triangles).