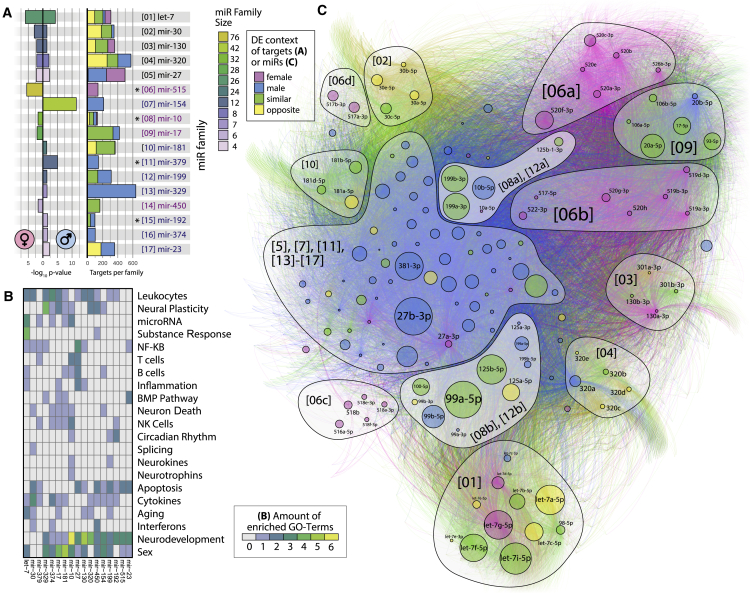
Figure 5.
Differentially Expressed miRNA Families Enriched in LA-N-2 and LA-N-5 Cells Self-Organize in a Comprehensive Targeting Network
(A) The p values of 17 enriched families (left) in LA-N-2 and LA-N-5 cells (female/male symbols) and mean predicted gene target count of each family (right). Color denotes family size (left) or contribution of sex to total target numbers (right), as determined via the DE context of individual family members. Asterisks indicate families with significantly smaller gene target sets (see text).
(B) GO topics curated for enriched families. Frequent terms are immune-, neurodevelopment-, or sex-related. The mir-10 and mir-199 families show rare association with neurokine signaling and circadian rhythm.
(C) The comprehensive network of all DE members of enriched families targeting 12,495 genes self-separates into family-dependent clusters by application of a force-directed algorithm (46,937 unique interactions). miRNA node size denotes absolute count-change, and color denotes DE context. Numbers in brackets correspond to (A). The mir-10 and mir-199 families form two distinct, sexually dimorphic clusters near the center of the network (lighter background color).