Figure 6.
The Cholinergic/Neurokine Interface and Experimental Validation of AChE Targeting by hsa-miR-125b
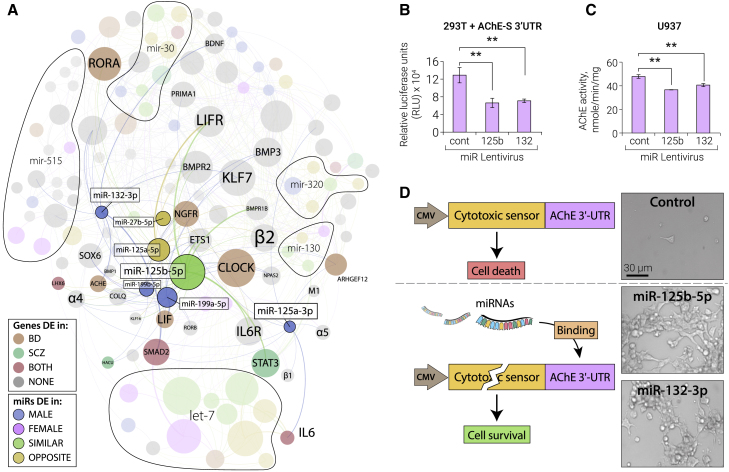
(A) The miRNA families mir-10 and mir-199 pose a sexually dimorphic interface of cholinergic, neurokine, and circadian regulation by targeting nicotinic/muscarinic (e.g., α4β2 and M1) and neurokine receptors, transcriptional regulators of cholinergic differentiation (LHX and STAT) and circadian rhythm (CLOCK and RORA), the AChE and the AChE linker proteins PRIMA1/COLQ, and high-affinity choline uptake (HACU). Members of mir-10/199 families, spike-in miR-132-3p, and their targeted genes are shown in color, and other miRNA families that passed the multiple filtering are indicated as areas. miRNA node size corresponds to count-change and gene node size to connectivity; color and thicker edges indicate the DE context and experimentally validated connections.
(B–D) Validation experiments of AChE targeting by miR-125b-5p, with miR-132-3p as a positive control.
(B) Lentiviral expression of miR-132 and miR-125b suppresses luciferase fused to the 3′ UTR of AChE in HEK293T cells. Error bars indicate SE.
(C) Lentiviral expression of miR-132 and miR-125b suppresses the endogenous AChE hydrolytic activity of U937 cells with similar efficacy. Error bars indicate SE.
(D) Life/death assay of stably transfected HEK293T cells carrying the AChE 3′ UTR fused to a cytotoxic sensor and co-transfected with miR-125b-5p, miR-132-3p, or control plasmids. Cells survive in case of binding of miR-132-3p and miR-125-5p to the 3′ UTR.