Figure 18.
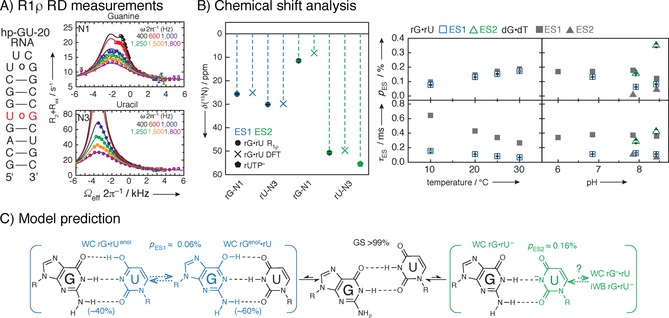
Example of an R 1ρ study of a 20‐mer RNA hairpin with GU base‐pair. A) Secondary structure of construct and 15N R 1ρ relaxation dispersion measurements with three‐state global fits of uracil N1 and guanine N3 of GU miss‐match base‐pair. Mono‐exponential decays with 4–20 points are recorded, and plotted against the offset referenced to the GS chemical shift, resulting in off‐resonance profiles. Fitting of >80 mono‐exponential and relaxation dispersion curves enables the detection of ES chemical shifts. B) Excited states can especially in the case of large chemical shift changes (>50 ppm) correspond to structures that have never been experimentally observed, as they are not a lowest energy structure. Therefore, methods other than structural database chemical shifts will have to be employed. In this case, to identify the underlying structures of the ES, structure‐based DFT prediction were employed. Population and lifetime of ESs as a function of temperature and pH was obtained to characterize the complex three‐state equilibrium. The chemical shift of ES1 and ES2 compared to structure‐based DFT prediction and rUTP ionization are shown in B. Population and lifetime of ES as a function of temperature at pH 6.9 and as function of pH at 20 °C indicated that detected ES1 represents rGNrUenol/rGenolNrU and ES2 ionized uridine, which was in agreement with excited states of GT base‐pair in DNA. C) Suggested exchange process depicts a multistate equilibrium between a GS wobble GU base‐pair and an ES Watson–Crick‐like GU base pair. Reprinted with permission from ref. 8.
