Figure 3.
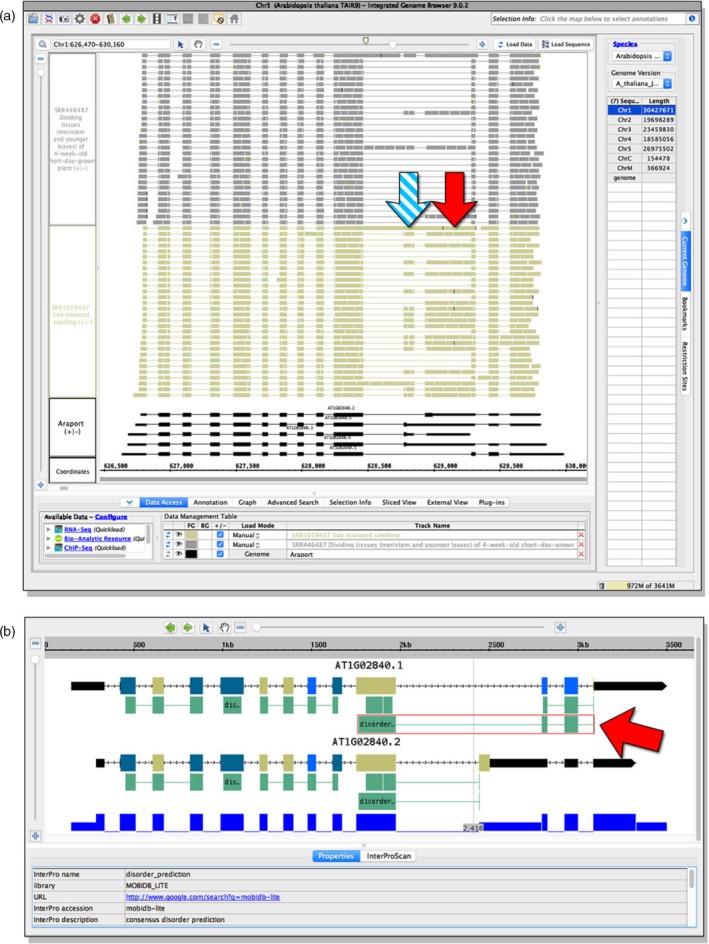
SR34 reads in dividing tissues (meristem and younger leaves) of 4‐week‐old short‐day‐grown plants and 5‐day‐old etiolated seedlings viewed with the Integrated Genome Browser (IGB).
(a) Dividing tissues and etiolated seedling sample reads visualized within the IGB main window. The solid red arrow highlights the reads within the etiolated seedling sample supportive of the alternative acceptor site for splice variant 2. The striped blue arrow highlights reads supporting an exon annotated as being in splice variant 3.
(b) The IGB ProtAnnot plug‐in app comparing results from HMMPfam and MobiDB‐lite for two SR34 splice variants, At1g02840.1 and At1g02840.2. The red arrow highlights the differing lengths of predicted disordered regions in the two splice variants.
