Figure 1.
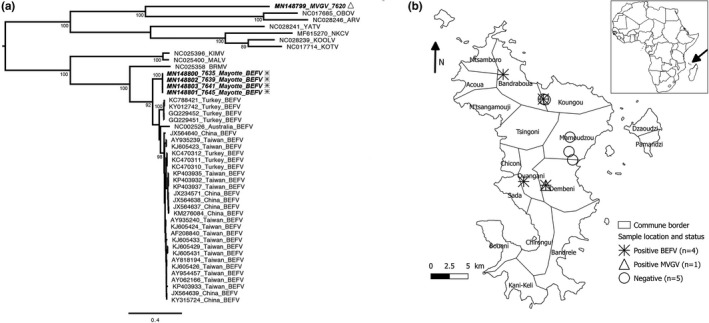
Phylogenetic analysis (a) and geographical location (b) and of the ephemeroviruses from Mayotte. (a) The tree was based on the full‐length glycoprotein nucleotide sequence and was constructed using the maximum‐likelihood approach based on the generalized time‐reversible model proportion of invariable sites plus gamma‐distributed rate heterogeneity (GTR+I+Γ4) utilizing SPR branch‐swapping, as estimated with PhyML 3.0 (Guindon et al., 2010). The robustness of individual nodes was estimated using 100 bootstrap replicates using a Bayesian‐like transformation of aLRT (aBayes). Only bootstrap values ≥ 90 are indicated. Scale bar indicates nucleotide substitutions per site. Ephemeroviruses from Mayotte are indicated in bold and in italic, with the same legend as (b). (b) The map of Mayotte shows the sample locations and their infected ephemerovirus status (the farm in Koungou reported two positive samples)
