Figure 1.
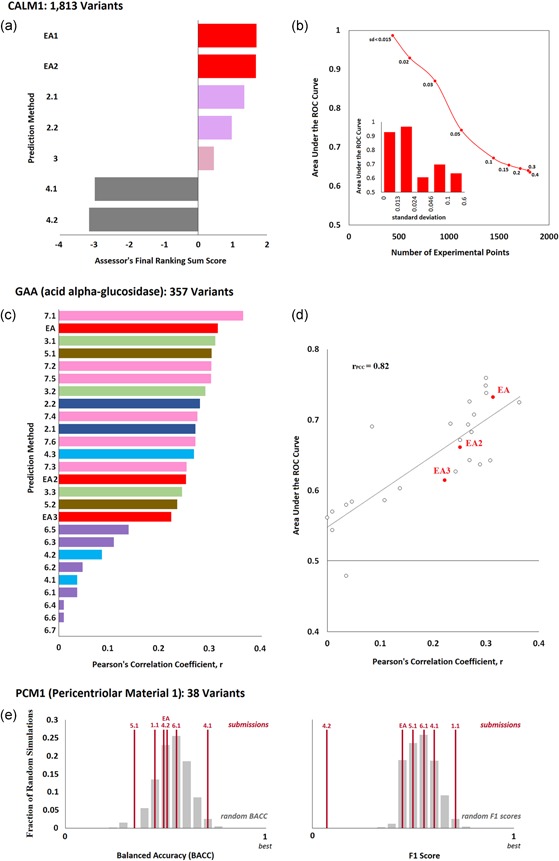
Effect of variants on protein function. (a) CALM1 challenge: Seven submissions aimed to predict the fitness effect of 1,813 variants of the human calmodulin measured with a competitive growth assay in yeast. The bar plot shows the final ranking sum scores for each submission, as calculated by the CAGI assessor. This score was derived from 16 different evaluation measures that represent three types of agreement (rank, original value, and rescaled value), and it is the sum of the average z‐scores of each type of agreement. Submissions from the same research team appear with the same color. (b) The area under the ROC curve (AUC) that corresponds to the Evolutionary Action submission (EA1) for data subsets defined by the experimental standard deviation values (SD). In the main panel, AUC was plotted as a function of the number of variants that have smaller SD than a maximum cutoff. The values next to each data point show the maximum standard deviation. In the insert bar plot, AUC was computed for five bins of variants that were created by sorting the variants according to their SD values and splitting them into nearly‐equal data point sets. (c) GAA challenge: 26 submissions aimed to predict the enzymatic activity of 357 variants of the human acid alpha‐glucosidase. The CAGI assessors used the Pearson's correlation coefficient (PCC) to assess the performance of the submissions. The bar plot presents the PCC for each method as calculated by the authors. (d) The area under the ROC curve versus the PCC values. The EA submissions are shown with red color. (e) PCM1 Challenge: Six submissions aimed to predict the effect of 38 human PCM1 variants on zebrafish brain development. The balanced accuracy (left plot) and F1 scores (right plot) of each submission are shown as vertical red lines, while the gray bars represent the corresponding distributions of 10,000 randomly generated predictions (calculated by the CAGI assessor). ROC, receiver operating characteristic
