Figure 4.
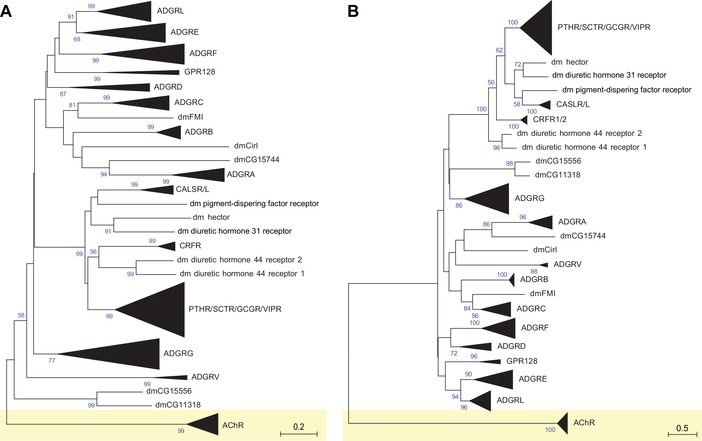
Phylogenetic relationship of Drosophila melanogaster aGPCR‐like sequences with vertebrate aGPCRs. The evolutionary relationships of D. melanogaster, human, mouse, chicken, and zebrafish aGPCRs are shown. Muscarinic acetylcholine receptors served as the outgroup. (A) The evolutionary history was inferred using the neighbor‐joining method.67 The optimal tree with the sum of branch lengths of 42.71172243 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches.68 The trees are drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method69 and are in the units of the number of amino acid substitutions per site. (B) The evolutionary history was inferred the maximum likelihood method based on the JTT matrix–based model.70 The tree with the highest log likelihood (–29511.43) is shown. Initial tree(s) for the heuristic search were obtained automatically by applying neighbor‐joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. The analyses involved 225 amino acid sequences. All positions with less than 95% site coverage were eliminated. That is, fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position. There was a total of 193 (A) and 141 (B) positions in the final dataset. Evolutionary analyses were conducted in MEGA7.22 dm, D. melanogaster.
