Figure 2.
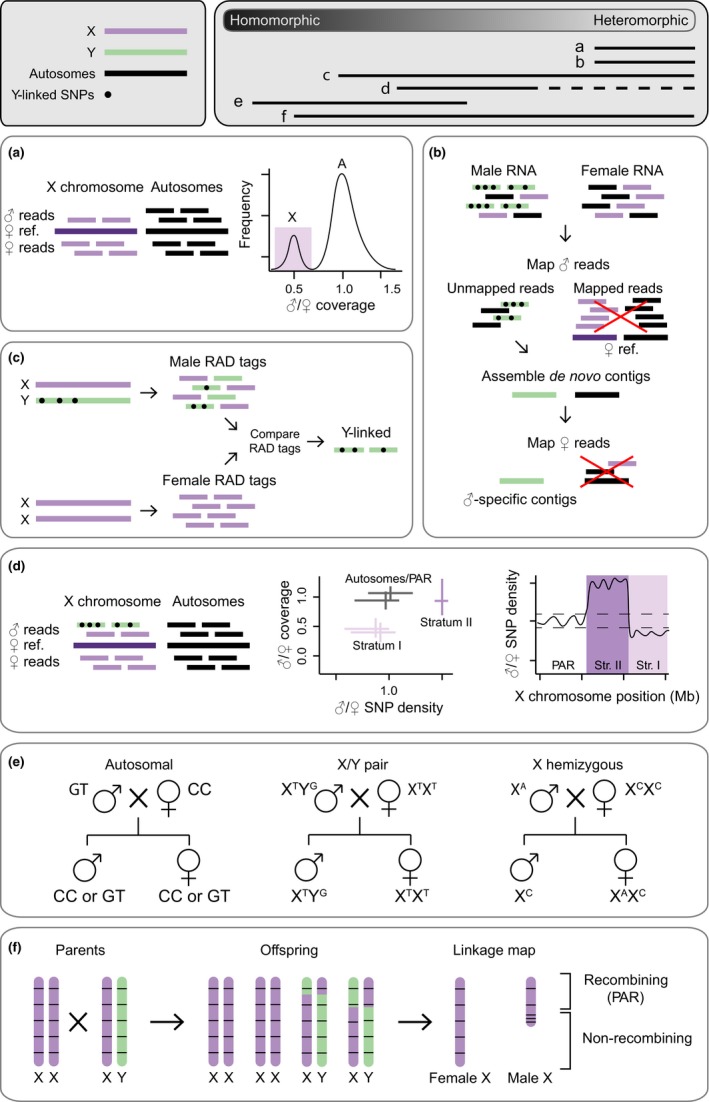
Overview of bioinformatic methods available for sex chromosome identification. This figure is based on XY sex chromosomes, but all methods can be inverted for ZW systems. Top left panel shows the key. Top right panel solid bars show which methods are most effective along different points of the sex chromosome divergence continuum. Dashed bar indicates that the method is partially effective. (a) Genomic coverage approach: in nonrecombining regions of sex chromosomes, where the Y has degenerated, males have only one X chromosome, and thus show a reduced genomic coverage relative to females. (b) Expression‐based approach: male RNA‐seq reads are mapped to a female reference. Unmapped reads are assembled into de novo contigs to identify putative Y‐linked sequences. Re‐mapping female transcripts to these contigs can be used to verify male‐limitation. (c) Association‐based approach: male and female RAD‐tags are compared to isolate male‐specific RAD loci. (d) SNP density approach: in younger regions of the sex chromosomes, which still retain high sequence similarity between the X and the Y, we expect an increase in male SNP density compared to females, as Y reads, carrying Y‐specific SNPs, still map to the homologous X regions. This SNP density pattern is not expected in old strata with substantial Y degeneration, as the X is effectively hemizygous in males. Contrasting sex differences in coverage and SNP density is a powerful approach to identify sex‐linked regions. (e) Segregation analysis approach: SNP data obtained from parents and progeny are analyzed in a statistical framework to assess the likelihood of autosomal versus sex‐linked segregation patterns. (f) Linkage mapping approach: recombination patterns of parents and offspring are compared, and regions with no recombination between males and females indicate putative sex‐linked regions
