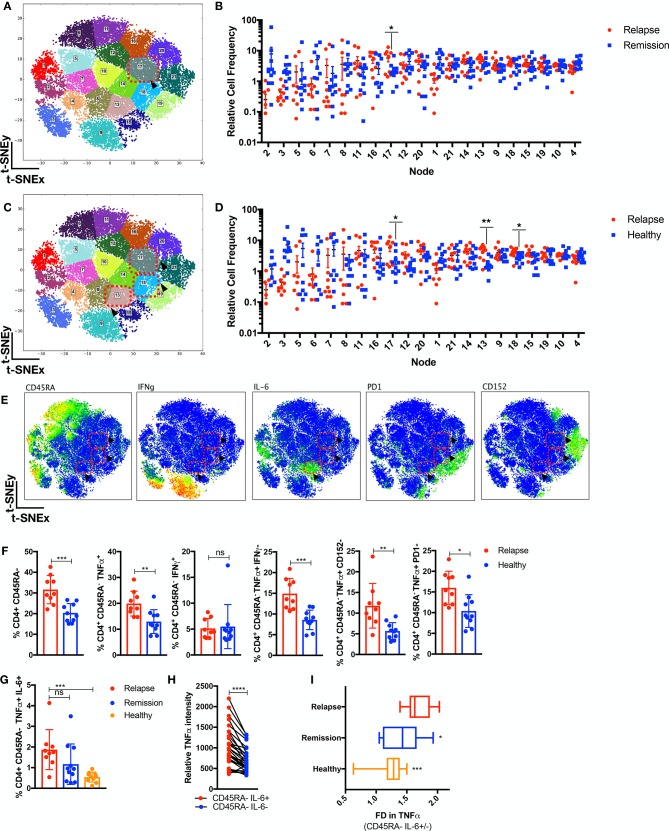
Figure 2.
Subclinical T-effector diversification in relapse individuals. CD4+TNFα+ cells from patients with JIA (n=20; relapse=9, remission=11) prior to therapy withdrawal and healthy paediatric controls (n=10) were analysed through t-SNE. (A) The CD4+TNFα+ t-SNE MAP is segregated into 21 nodes with enrichment in (B) node 17 (p<0.05) for relapse as compared with remission individuals and (C,D) node 17, 18 and 13 (p<0.05) for relapse as compared with healthy individuals. (E) The phenotype expression is shown for node 17, 18 and 13 (red-dotted boxes and arrowheads). (F) Supervised gating of preclustering FCS files validates the relevant CD4 memory cellular subsets in relapse versus healthy individuals. (G) Supervised gating of CD4+CD45RA−TNFα+IL-6+ cells in relapse/remission/healthy individuals. (H) Relative TNFα intensity in CD45RA−IL-6+ or CD45RA−IL-6− cells. (I) Fold difference (FD) in TNFα intensity in CD45RA−IL-6+ over CD45RA−IL-6− cells in relapse/remission/healthy individuals. Comparison of cellular subsets performed with Mann-Whitney U, unpaired or paired two-tailed test, means±SD. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. IFN, interferon; IL, interleukin; ns, not significant; TNFα, tumour necrosis factor alpha; t-SNE, t-distributed stochastic neighbour embedding.