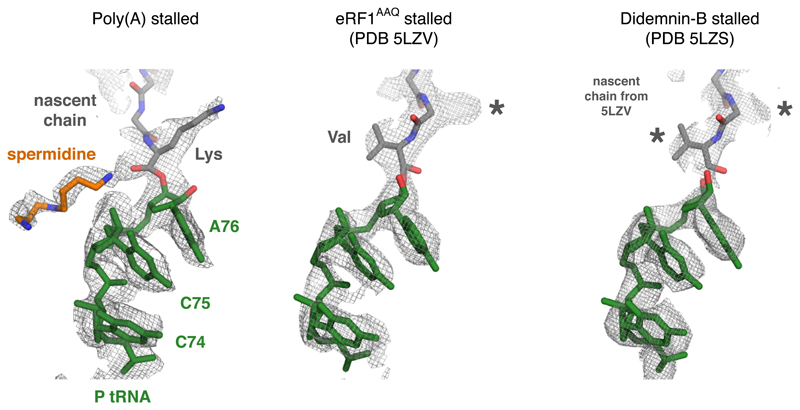
Extended Data Fig. 7. Comparison of peptidyl-tRNA geometry in different mammalian RNC structures.
Shown are the EM density maps for the peptidyl-tRNA region at the PTC for the indicated structures. The fitted models are shown for the poly(A)-stalled ribosome and the RNC stalled at the stop codon with a dominant-negative eRF1AAQ mutant (PDB code 5LZV). The 5LZV RNC is in a geometry competent for peptidyl-transfer (or in this case, peptide release by eRF1). The structure from the didemnin-B stalled RNCs contains a mixture of nascent chains stalled at different positions. Thus, the nascent chain density represents an average of a variety of peptidyl-tRNAs. Note that the nascent chain model from 5LZV fits well into the density map, indicating that the majority of peptidyl-tRNAs assume this configuration during active elongation. The geometry for the poly(A) peptidyl-tRNA is unambiguously different from this optimal geometry. Lys and Val refer to the lysine and valine side chains of modeled nascent chains. The asterisks indicate density for side chains that are not shown.