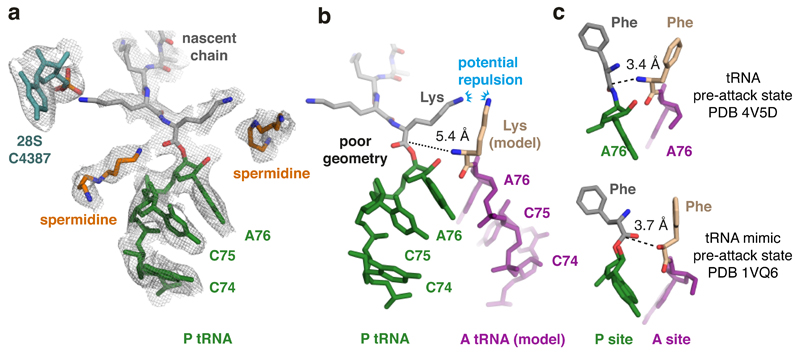
Fig. 4. Peptidyl-tRNA is mis-positioned in the poly(A) stalled ribosome.
(A) The P-site tRNA (green), attached nascent chain with the first three lysines (grey), and 28S rRNA residue C4387, which interacts with the penultimate Lys side chain, are shown fitted within the EM density map (mesh). Putative spermidine molecules (orange), hundreds of which are thought to bind ribosomes and facilitate translation50,51, were modeled into otherwise unaccounted density. (B) Superimposed models of the peptidyl-tRNA in the poly(A)-stalled ribosome with the A-site tRNA positioned for peptidyl transfer (PDB 4V5D). The amino acid in this structure (phenylalanine) was replaced with lysine to model the situation during poly(A) translation. The proximal lysine of the nascent chain faces the lysine of the accommodating A-site tRNA, resulting in potential charge repulsion between their respective epsilon amines. The backbone geometry of the nascent chain attachment to A76 is suboptimal for peptidyl transfer, as highlighted by the 5.4 Å distance between the α-amino group of the aminoacyl tRNA and the incorrectly oriented backbone carbonyl of the peptidyl tRNA. (C) Shown are models depicting the P- and A-site amino acids attached to the P- and A-site nucleotide (A76) for the indicated structures. Note that in both pre-attack structures, the attacking atom is within 4 Å of the P-site target bond, unlike the 5.4 Å distance in the poly(A)-stalled ribosome.