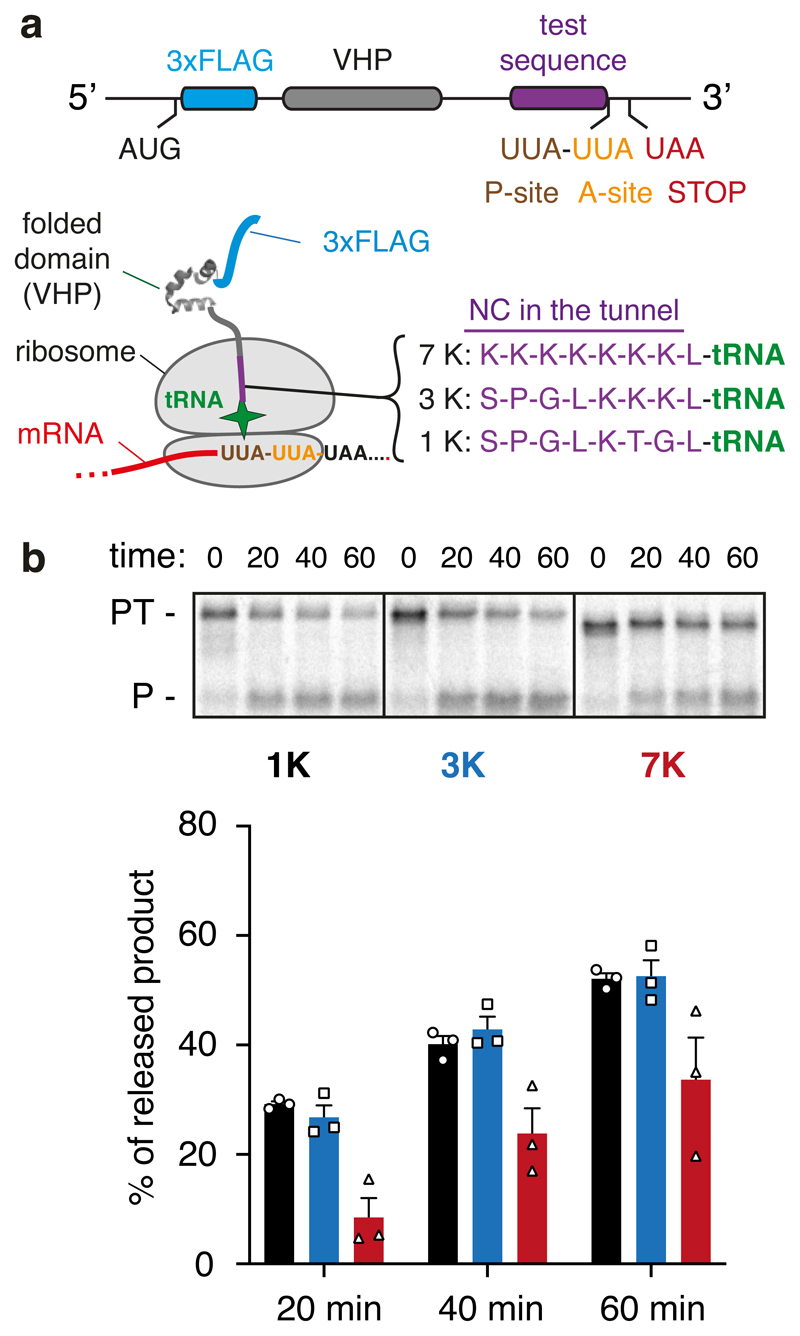
Fig. 5. Analysis of PTC geometry by puromycin reactivity.
(A) Experimental strategy. A construct was designed to cause ribosome stalling at the second of two rare UUA leucine codons (see Methods). This RNC is known to be functional because translation resumes when liver tRNA is added53. RNCs were produced using this strategy in which one of three test sequences (7K, 3K, and 1K, as indicated) are positioned inside the ribosome tunnel. The lysine residues were encoded with AAG codons to minimize stalling before the desired UUA codon. After the translation reaction to synchronise ribosomes at the rare codon stall, the sample was moved to ice and the salt concentration was increased to 500 mM to prevent splitting of ribosomal subunits by rescue factors. Puromycin was then added to 2 μM final concentration and polypeptide release from tRNA was judged by SDS-PAGE. (B) Stalled RNCs containing the 1K, 3K, and 7K test sequences were evaluated for their reactivity to puromycin over a 60 min time course as a measure of peptidyl-transfer capacity. A representative experiment out of three is shown in the top panel, with the bottom panel showing a graph depicting the mean ± SEM (n=3) and individual data points from independent experiments. The positions of peptidyl-tRNA (PT) and puromycin-released peptide (P) are indicated to the left of the gel. Uncropped gel image is available online (Source Data).